Updates to 3D Slicer Volumetric Mesh Support

In a previous blog post, the addition of volumetric mesh support to 3D Slicer was announced. In this post, we report two new features to better visualize 3D volumetric meshes: 1) An integrated model clipping method for both surface and volumetric meshes and 2) Opacity control for 2D slice intersections of models.
Integrated model clipping method
When volumetric mesh support was added to 3D Slicer, volumetric and surfaces meshes were clipped using two distinct methods. Surface meshes were clipped to a smooth surface (i.e. the displayed mesh would contain “partial” cells from the source mesh), while volumetric meshes were clipped to only display whole cells. The recent update allows both mesh types to be clipped with either method using an option in the Models module, and provides an additional clipping option available through the API.
The ClippingTypeMethod member variable has been added to vtkMRMLClipModelsNode to control the selection of different clipping methods. The MRML node currently supports three clipping options: Straight (matches clipping plane exactly, leaving partial cells in view); Whole cells (boundary cells that cross the clipping plane are removed); and whole cell including boundary cells (boundary cells are preserved). Straight clipping is implemented using vtkClipPolyData and vtkClipDataSet for surface and volumetric meshes, respectively. Whole cell extraction clipping is implemented using vtkExtractPolyDataGeometry and vtkExtractGeometry. The source changes for the update can be found here.


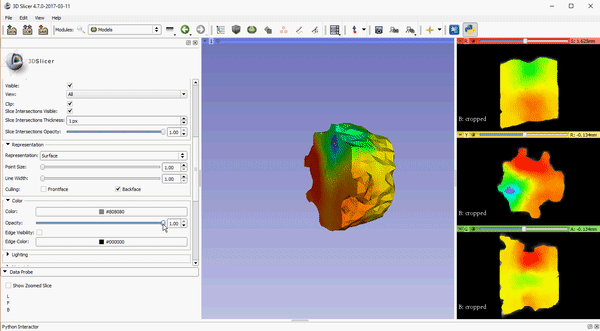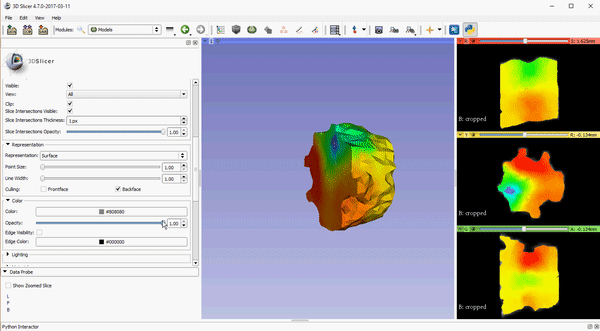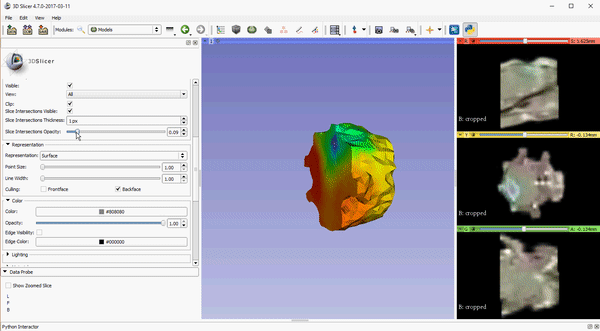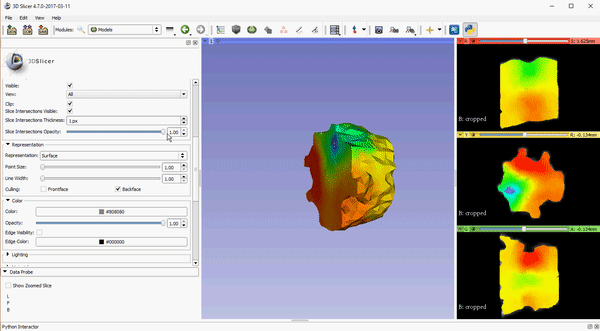
Opacity control for 2D slice intersection in models
To improve visualization of volumetric meshes alongside volume data, independent control of the opacity of 2D slice intersections has been added to the Models module. The source changes for the update can be found here.
These features, which are already available in the Slicer Nightly build, will be a part of the 4.7 release of 3D Slicer. To download the software, please visit http://download.slicer.org/. To learn more about how Kitware can tailor solutions to meet your research needs, please contact kitware@kitware.com.
Acknowledgments
This work was supported by the National Cancer Institute under award number R01CA184354 as part of NIRFAST-Slicer improvements for NIRFAST. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.