slicerSMTK: Bridging CMB and 3D Slicer
Last year, Kitware announced the aeva (annotation and exchange platform for virtual anatomy) application, which aims to provide a streamlined interface for the generation, modification, annotation, and exchange of anatomy in medical images/scans. It supports diverse data formats and standards. As part of aeva 2.4, we have released a slicer extension module (slicerSMTK) that helps bridge CMB and 3D Slicer. This extension module will help streamline the usage of aevaSlicer and aevaCMB by facilitating the transfer of data between the two applications.
Background
The aeva application suite contains two applications: aevaSlicer and aevaCMB. aevaSlicer, which is based on 3D Slicer, is designed to provide comprehensive image segmentation, robust surface geometry generation, and a multitude of volumetric meshing capabilities. Whereas aevaCMB – which is based on CMB (Computational Model Builder), SMTK (the Simulation Modeling Tool Kit), and ParaView – provides resources and templates to edit, subset, and mark up features of unstructured geometric data with ontological terms (from anatomical ontologies such organ and tissue type; as well as other ontologies).
Previously, simulation and modeling engineers using the aeva suite had to manually select and load imaging and modeling data individually to aevaCMB after generating them in aevaSlicer. To simplify this process and make the two applications more interoperable, we built an extension module called slicerSMTK and released it as part of aeva version 2.4.
slicerSMTK extension module
At the core of CMB is a lightweight library known as Simulation Modeling Toolkit (SMTK). This library provides an interface for representing geometric models and meshes, the operations that can be performed on the models and meshes, and complex information structures. SMTK is useful to associate information with various components of the geometric domain in order to define a complete and consistent annotated model (for simulations and other purposes).
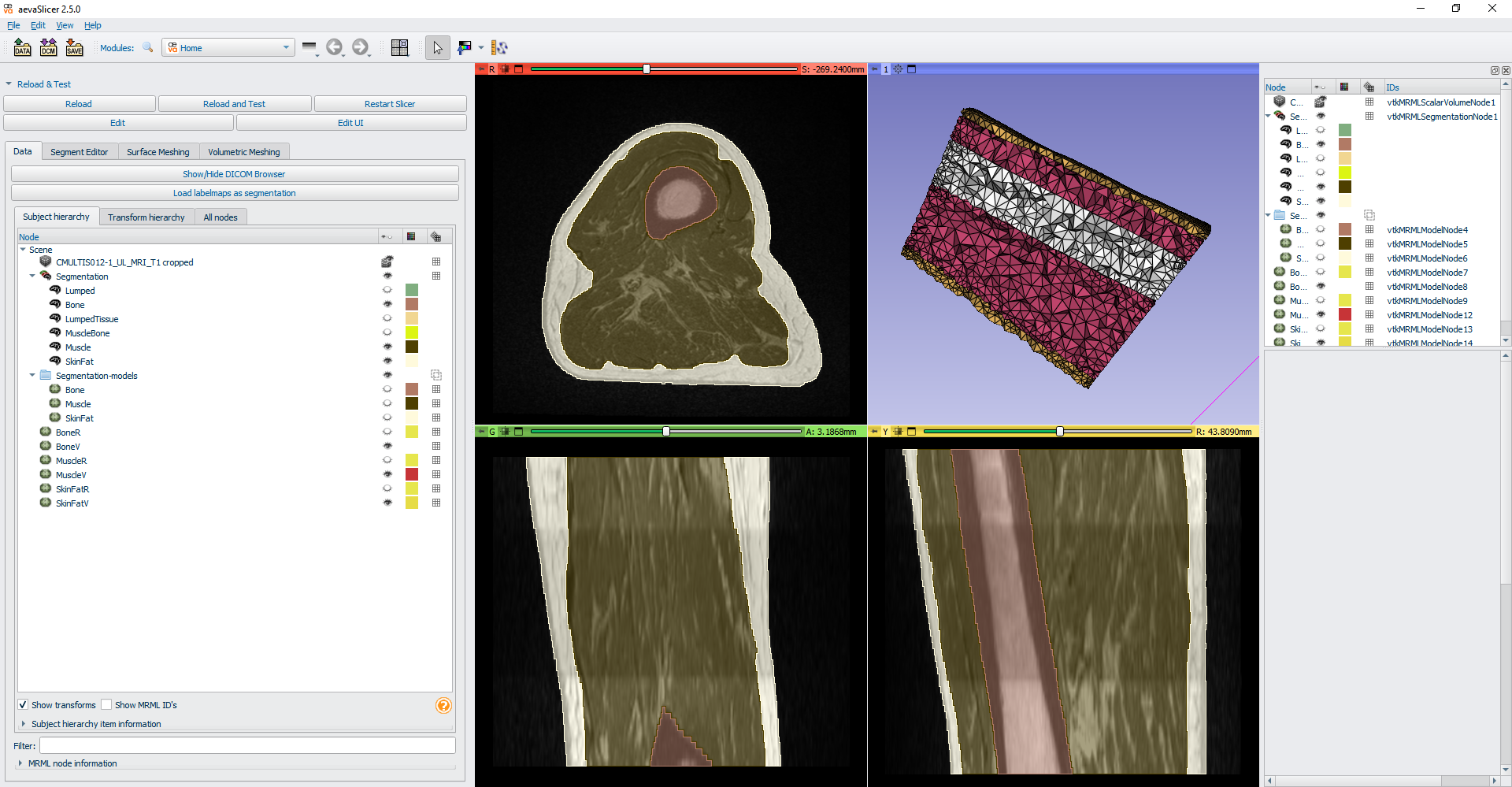
The slicerSMTK extension module allows the 3D Slicer’s MRML scene to be exported as an smtk file set. As shown in the figure below, “aeva.smtk” is one of the file extensions that you can write out as a 3D Slicer scene file.
https://gitlab.kitware.com/aeva/slicersmtk
The exported smtk data consists of a smtk json file with data description and organization and a folder that contains the image and model data in vti and vtp formats/.
Aeva 2.4 installers can be found here and the slicerSMTK extension module source code can be found here.
Using slicerSMTK
Implementation detail
The exported JSON file (.aeva.smtk) stores the high-level scene information associated with different models and images in the scene, along with URLs pointing to the individual data files. The extension exports surface meshes as vtkPolyData (.vtp), volume meshes as vtkUnstructuredGrid (.vtu), and images/volumes as vtkImageData (.vti). Individual component files (vtp, vtu, vti) are saved under a subfolder with the same name as the scene file. The components undergo transformation hardening before the export operation, thereby carrying forward any registrations/transformations applied within Slicer. Names of individual component files are made more readable by using the user-provided names for the corresponding MRML nodes as a prefix to the file followed by their respective uuid. The prefix undergoes string sanitization to ensure that it is compatible as a file name.
Transformation hardening and filename sanitization are core functionalities provided by 3D Slicer, whereas the functionality to export SMTK files is provided by aeva-session which is an extension of SMTK and also acts as a ParaView plugin used by aevaCMB.
Currently, the extension only builds as a module bundled with aevaSlicer. Future releases will include standalone/external building capability as well as reading/loading back the smtk file into the Slicer application.
To learn more about the CMB and 3D Slicer platforms and how we can help you build a simulation and modeling custom application, please contact us at kitware@kitware.com.
Acknowledgment
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB025212. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.