SlicerRVXLiverSegmentation: 3D Slicer extension for interactive liver anatomy segmentation


Annotation plays a key role in clinical practice and in the creation of reference datasets that are useful to evaluate medical image processing algorithms and to train machine learning based architectures. Since softwares embedded into acquisition machines are not always accurate for this task, researchers and engineers have developed more and more interactive annotation and segmentation tools. But, despite these developments, annotated datasets are still missing for many clinical applications.
The creation of the SlicerRVXLiverSegmentation extension has been motivated by the dramatic absence of a public dataset providing hepatic MRI data together with their ground-truth segmentations of liver anatomy. By creating our own dataset, we have developed a plug-in specifically dedicated to this task, that can be used publicly by any user for the construction of new datasets related to liver pathologies. Moreover, our plug-in has numerous clinical applications, such as the hepatic volumetry, which is becoming increasingly important in view of the growing number of chronic hepatopathies requiring transplants or hepatectomies.
About the extension
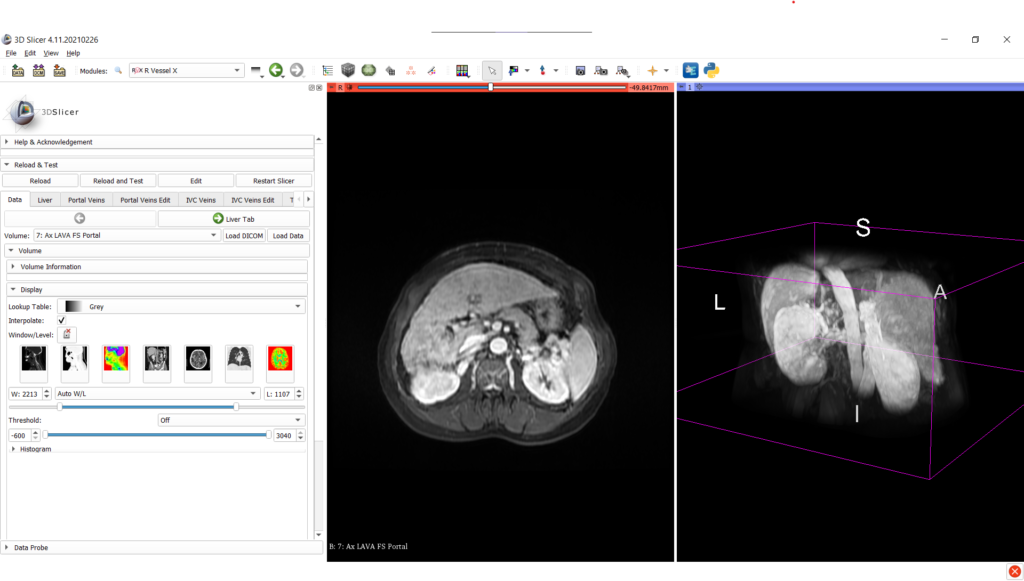
For research purpose needing annotation of liver anatomy from medical images, the SlicerRVXLiverSegmentation provides 7 main tabs:
- loading and managing medical imaging data;
- liver segmentation;
- annotation of portal veins and segmentation;
- editing portal veins segmentation;
- annotation of inferior vena cava and segmentation;
- editing inferior vena cava segmentation;
- tumor segmentation.
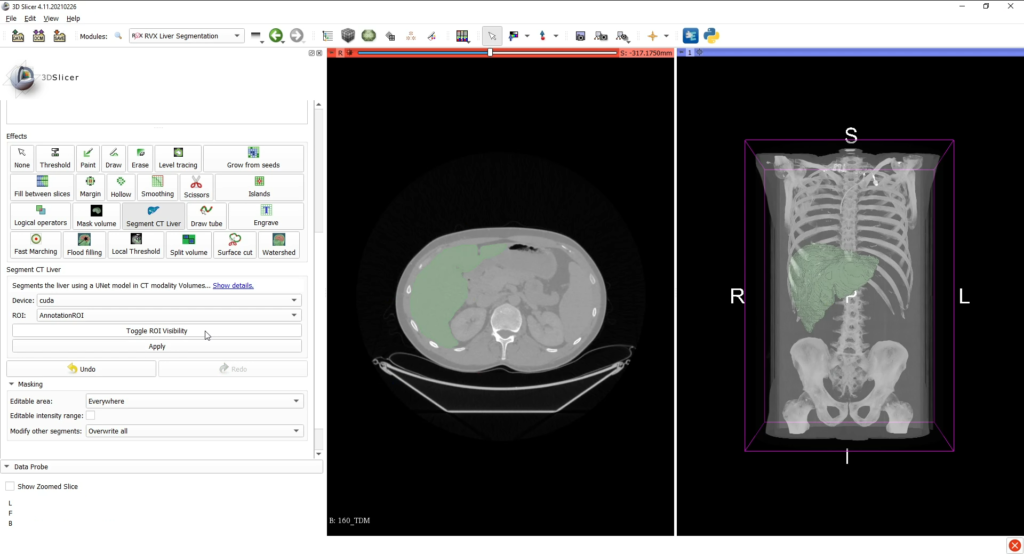
Once the medical image data is loaded into the 3D Slicer interface, the liver can be segmented with the associated tab, either by using interactive tools (such as region growing approaches) or by an automatic deep learning (for CT scans only).
The AI CT segmentation is directly integrated into 3D Slicer’s segment editor as a a segment editor effect. The model was developped using the MONAI framework and was trained on the Medical Decathlon Dataset.
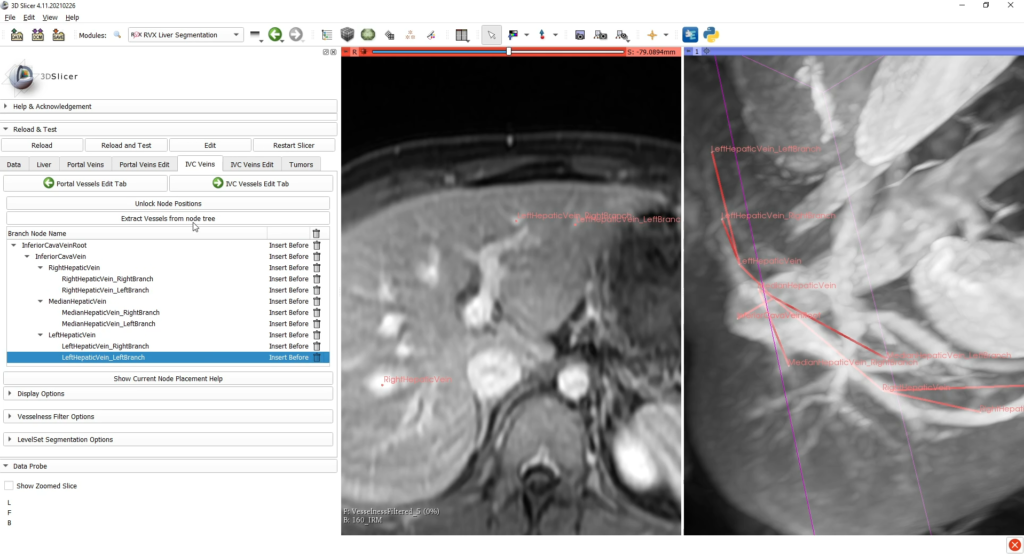
Then, the reconstructions of hepatic vessels (portal vein and inferior vena cava) are based on tree structures interactively built by the user, who places the nodes of important branches and bifurcations (with specific anatomical nomenclature) into the scene of the medical image to be processed.
After this step, a VMTK (Vascular Modeling Tool Kit) module segments the vessels by using those graphs as initialization patterns; also, the user can verify and edit this segmentation. The last tab allows the user to segment interactively possible tumoral tissues with dedicated tools.
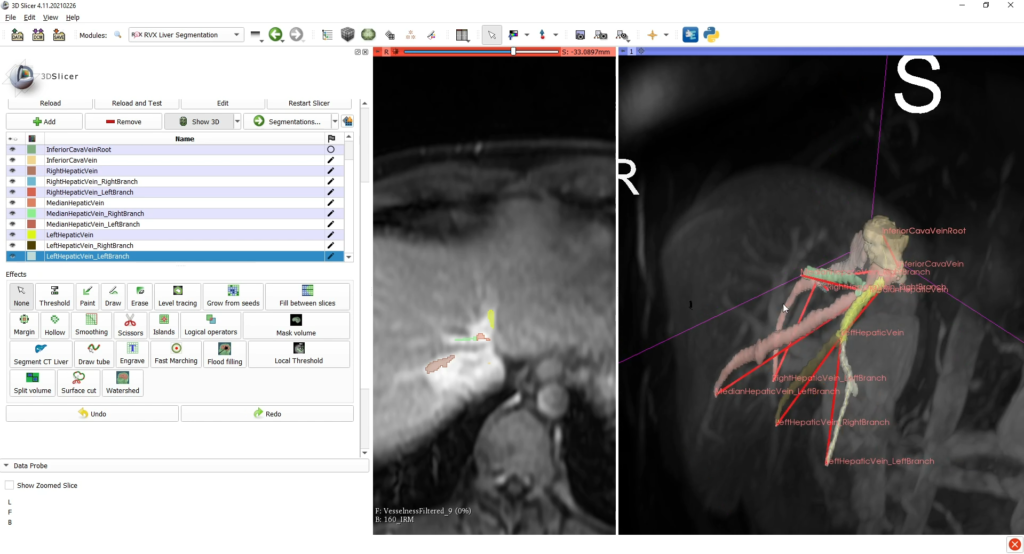
This tab also permits to export the complete scene, comprising:
- segmentation label maps (liver, inferior vena cava, portal vein, tumors);
- portal vein and inferior vena cava intersection positions (fiducial CSV and adjacency matrix);
- the complete scene as a MRB file.
Future works
We first would like to integrate advanced deep learning models for liver and hepatic vessels segmentation into our SlicerRVXLiverSegmentation plug-in, in order to provide automatic reconstructions that can be then edited by the user with the other tools proposed in the plug-in and in 3D Slicer.
Another important work concerns the VMTK module, which needs more adaptations for MRI processing. As an example, the Frangi filter is employed as a pre-processing step (as a vascular enhancement algorithm), while other approaches could be opted instead.
Finally, a more complete evaluation protocol will be conducted with our plug-in, compared to commercial solutions, by taking into account larger patient cohorts.
Execution
Kitware’s partners on this project are the LIRIS laboratory, Pascal institut, CReSTIC laboratory, and the CHU Clermont Ferrand.
This work was funded by the French Agence Nationale de la Recherche (grant ANR-18-CE45-0018, project R-Vessel-X, http://tgi.ip.uca.fr/r-vessel-x). Antoine Vacavant is coordinator of the project (CHU Clermont Ferrand, Pascal institut).