Quantifying Anatomical Shape with SlicerSALT

Two years ago, the National Institute of Biomedical Imaging and Bioengineering funded an initiative to create open source software to enable biomedical researchers to generate shape analysis measurements from their medical images. This software is called Slicer Shape AnaLysis Toolbox (SlicerSALT). It is designed to re-engineer state-of-the-art shape modeling and analysis research code into a robust, bundled toolkit. More specifically, SlicerSALT aims to enhance the intuitiveness and ease of use of research code for shape analysis studies. It also aims to help researchers find changes in shape with higher statistical power.
This article demonstrates how to use SlicerSALT to compute a spherical harmonic representation of a random object of spherical topology (the Stanford bunny). The data for the example is available through the link below, and SlicerSALT is available as source code and as binary distributions for all major platforms (e.g., Linux Redhat/CentOS, Windows and MacOS). Links to the source code and distributions reside on salt.slicer.org.
Example Data: http://bit.ly/kwsource-slicersalt-example-data
Stanford Bunnies
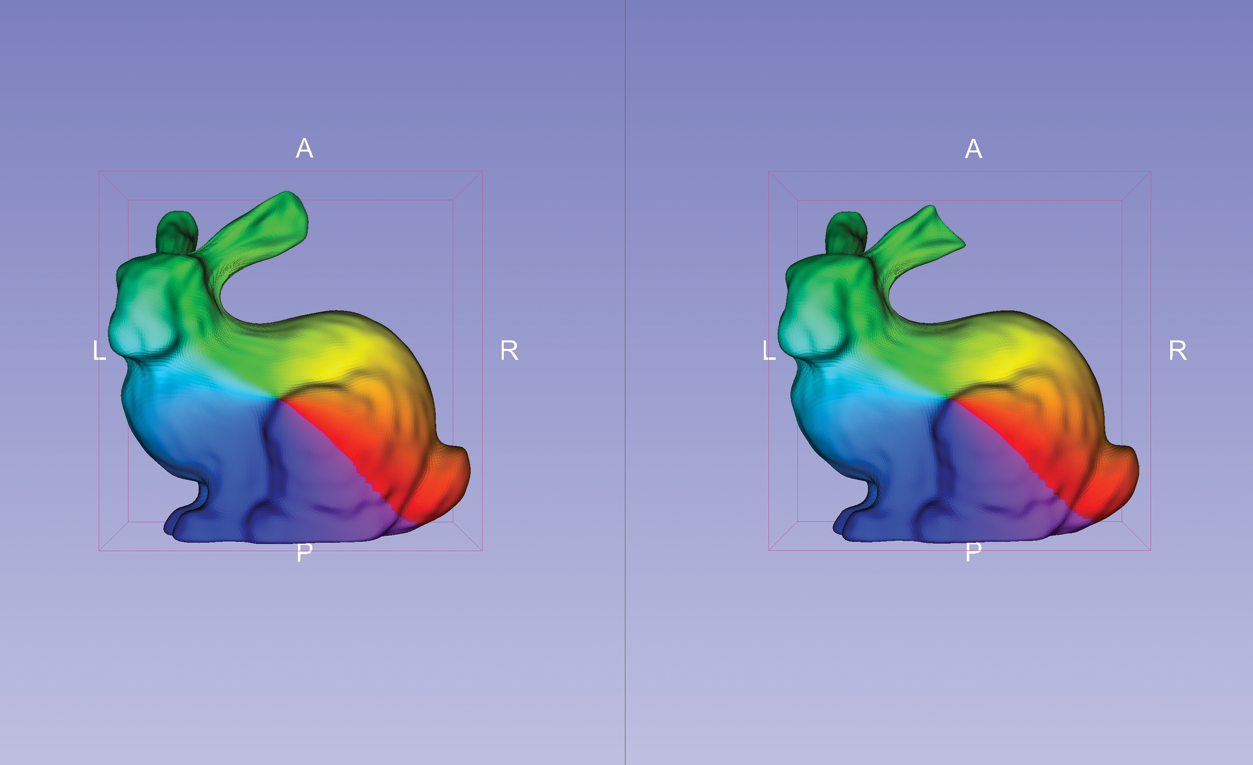
SlicerSALT’s Shape Analysis Module densely samples landmarks based on the geometric properties of spherical shapes. It was used to create the screenshot to the right that shows corresponding meshes of two Stanford bunnies: an original and another with a cut ear.
Shape Analysis Module contains SPherical HARmonics-Point Distribution Model (SPHARM-PDM). SPHARM-PDM is a hierarchical, global, multi-scale boundary description that can only represent objects of spherical topology that are based in spherical harmonics. SPHARM-PDM
can be computed from binary segmentations
or surfaces.
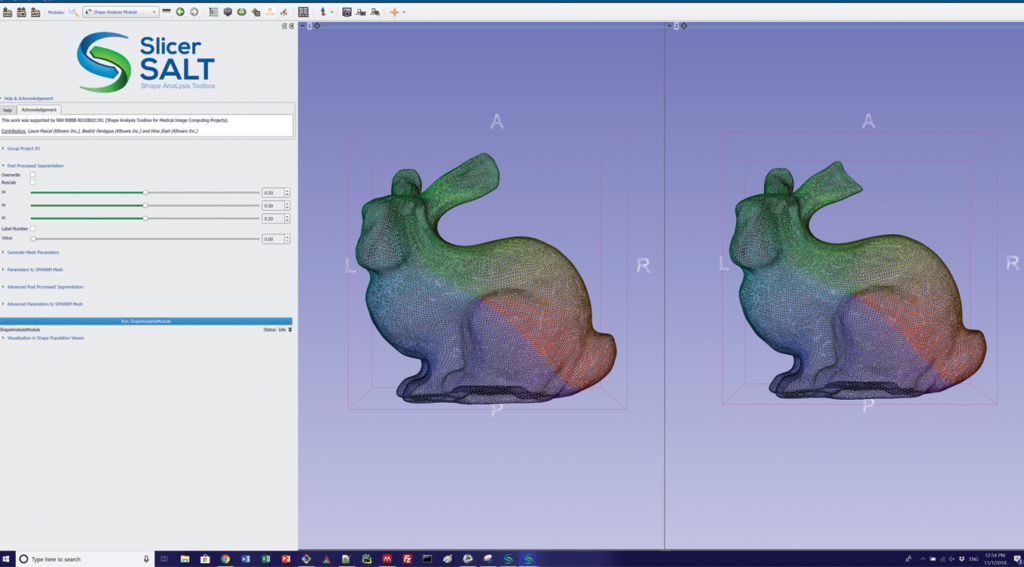
In SlicerSALT, the binary segmentations or surfaces are pre-processed by a filter to ensure they have spherical topology. More specifically, the filter smoothes and fills small interior holes. The pre-processed inputs are then converted to quadrilateral surface meshes. From the meshes, SlicerSALT computes spherical parameterization with area-preserving, distortion minimizing spherical mapping. SlicerSALT implements this mapping to fit a spherical harmonics basis, which it uses to compute a SPHARM description for each input dataset. SlicerSALT then samples the SPHARM description to generate triangulated surfaces (or SPHARM-PDM representations) via icosahedron subdivision. These representations have geometric correspondence between structures, as they contain analogous points in similar parts of their geometry.
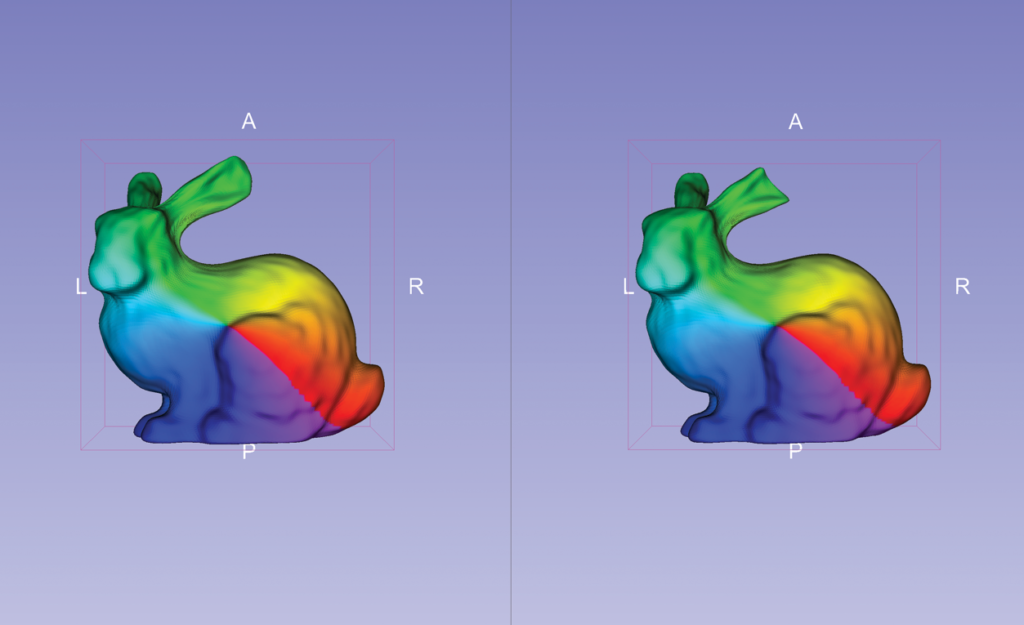
In medical applications, SPHARM-PDM representations of the same anatomical structure can be used to quantify shape differences. SlicerSALT, for example, can use such representations to quantify a defect of a known magnitude between two models. In the case of the Stanford bunnies, the defect is a cut ear.
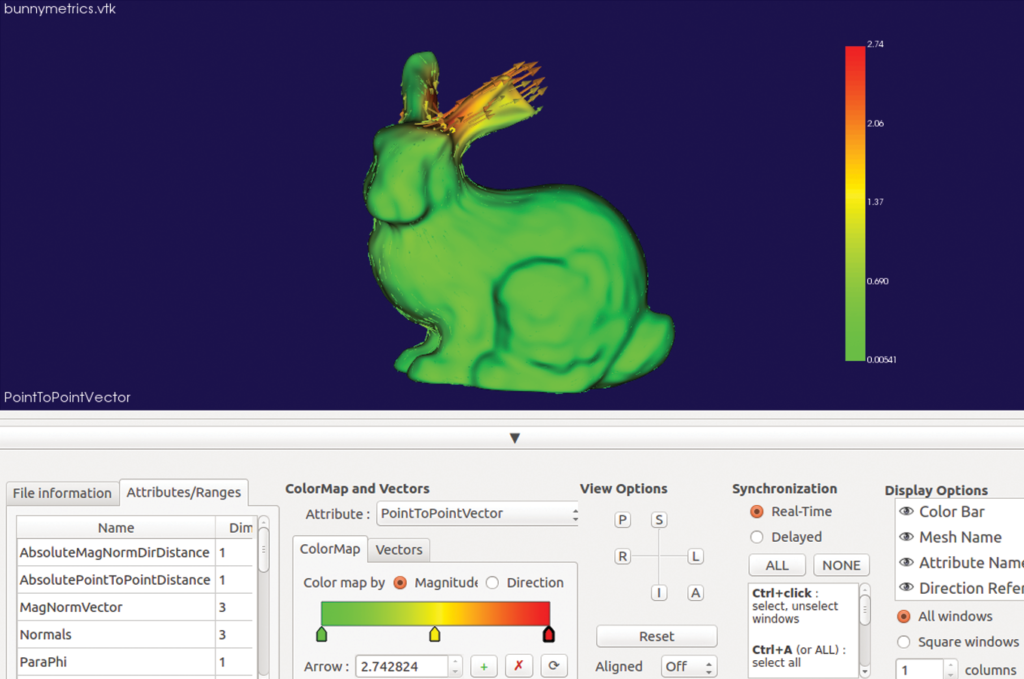
In particular, two modules in SlicerSALT compute and visualize point-to-point correspondent vectors that quantify the localized differences between shapes. These modules are Model to Model Distance and Shape Population Viewer.
For the Stanford bunnies example, SlicerSALT finds that the ear of one bunny was cut roughly three millimeters. This simple example illustrates the precision of SlicerSALT’s quantification methods, which can be used to measure shape defects in anatomy in the presence of disease or treatment.
Acknowledgement
Research reported in this publication was supported by the National Institute Of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01EB021391. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

Beatriz Paniagua is a technical leader on the medical computing team at Kitware, where she leads craniomaxillofacial and musculoskeletal image analysis projects as well as morphometry analysis projects. Her main research interests are computational anatomy and the translational aspects of science.
Dear Beatriz Paniagua, I’m Kaiwen, a graduate student in medical school. I am very interested in the SPHARM-PDM. I have used the Example Data mentioned in this paper for the SPHARM-PDM analysis. I found that the original two Bunnies overlapped well, but the position where the two Stanford Bunnies could not overlap seemed to increase after the SPHARM-PDM calculation. These are my question:
1. Will the SPHARM-PDM calculation lead to the displacement of the two models? Does the displacement affect the shape defects analysis of the Stanford Bunnies?
2. If I want to calculate the corresponding point distance between the two models after the calculation of the SPHARM-PDM, is it necessary for me to register the models after the calculation of the SPHARM-PDM, and then calculate the corresponding point distance between the models? If so, which registration should I choose?
I’m looking forward to your reply.