Project Spotlight: DARPA POCUS
Revolutionizing Trauma Care with AI Solutions
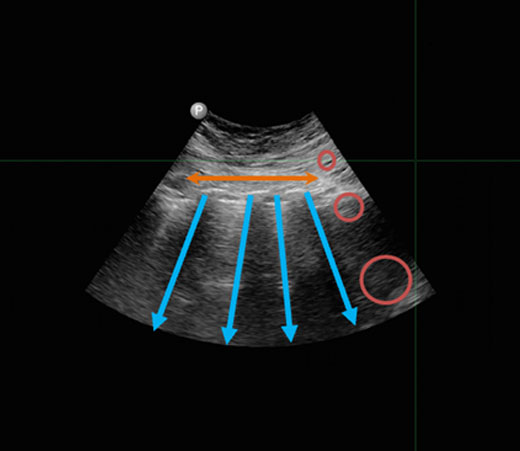
Ultrasound imaging is a valuable medical tool that helps physicians evaluate, diagnose, and treat various conditions quickly. While ultrasound devices have become more portable, having a physician with the appropriate expertise on-site at difficult and dangerous locations (e.g. battlefields) is not always feasible. Through our work on DARPA’s Point-Of-Care Ultrasound Automated Interpretation (POCUS AI) program, we’ve trained artificial intelligence (AI) to mimic the image interpretation processes of Point-Of-Care Ultrasound (POCUS) experts.
The Challenge
Deploying portable POCUS devices to the battlefield is crucial for quickly and accurately addressing a wide range of injuries. However, several challenges hinder the deployment of this technology, including logistical issues, financial constraints, and—most critically—the lack of frontline medical personnel trained to use these instruments. DARPA POCUS aims to overcome these challenges by advancing artificial intelligence (AI) techniques that interpret ultrasound videos, enabling medical personnel to make informed decisions even in remote and resource-limited environments. Additionally, to reduce the high costs associated with developing medical imaging AI, the program is pioneering new methods that require only tens of ultrasound videos for training, as opposed to the thousands typically needed. This significant reduction in training data is achieved by leveraging the expertise and insights of current ultrasound professionals.
Leveraging Open Source Tools for Medical Imaging AI
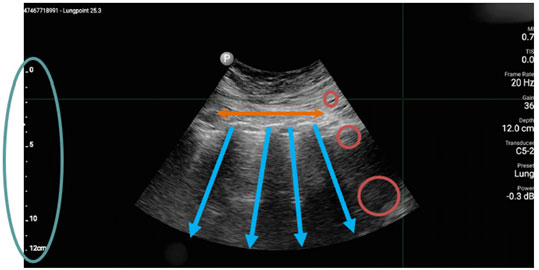
Kitware was selected to participate in Phase 1 of this project. Our team included Bharadwaj Ravichandran, Christopher Funk, Ph.D., Brad Moore, Ph.D., Jon Crall, Ph.D., and Stephen Aylward, Ph.D., and we partnered with with Sean Montgomery, M.D. and Yuriy Bronshteyn, M.D. from Duke Hospitals. Our team focused on utilizing three open source tools: MONAI, ITK, and KitwareMedical/ImageViewer. MONAI, an open source platform designed to address the challenges of integrating deep learning into healthcare solutions, provided GPU-optimized pipelines that incorporated ITK filters for data transformation and augmentation. This allowed us to compare multiple AI network architectures and training strategies, providing best-in-class solutions and cutting-edge algorithms for medical imaging AI. MONAI also offered explainable and ethical AI methods to explore our approach’s strengths and weaknesses, facilitate insightful discussions with clinicians, and identify promising paths for future work.
The next tool we leveraged is ITK, an extensive suite of software tools used for image analysis. Consistent feature appearance in ultrasound images can be affected by the varying orientation of transducer lines across an image. We developed an ITK filter to overcome these inconsistencies using a novel transducer line vs. depth mapping. This filter estimates the path of each transducer line and then encodes the information along each line based on its depth from the skin surface. This approach preserves the linear properties of the space while spreading out the information compressed near the probe.
Lastly, we enhanced the KitwareMedical/ImageViewer to significantly speed up the 2D+time image labeling process, resulting in improved labeling accuracy. We increased the annotation speed by implementing a binary morphological interpolation scheme to interpolate labels between annotated slices. This allowed the medical researchers to annotate only when significant movements or changes in shape occur in the objects of interest. The interpolation algorithm fills in the annotations for the frames in between these sparse annotations, significantly speeding up and improving the accuracy of training data generation.
Ethical AI and Medical Imaging
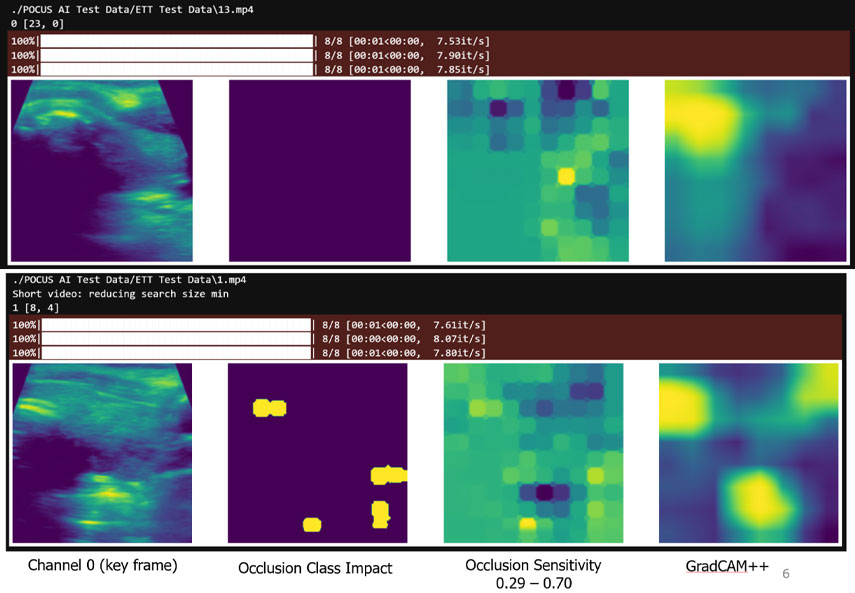
Bottom row: ETT misidentification occurs when multiple extraneous locations degrade the localization. Those extraneous locations can only be distinguished in the video by considering a larger context.
To gain insight into the AI’s decision-making process when interpreting ultrasound images, we used MONAI’s explainable and ethical AI computation and visualization methods. For example, for the endotracheal tube (ETT) identification task, we used MONAI’s implementation of Occlusion Sensitivity and GradCAM++ to study correctly and incorrectly identified cases. These studies revealed that while our network almost always identified the proper location of the tube, it sometimes was confused by other, similar-looking structures in the videos. Our experts confirmed that a larger context is needed to differentiate these structures, so transformer networks are now being investigated as they specifically support larger contexts.
Results and Impact
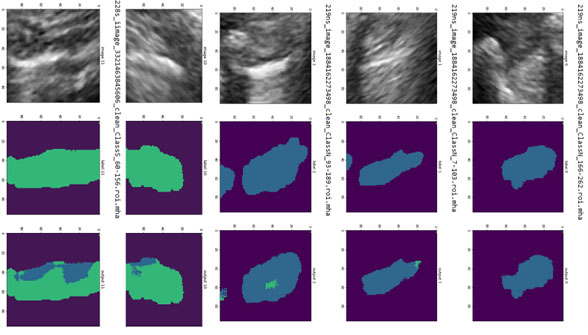
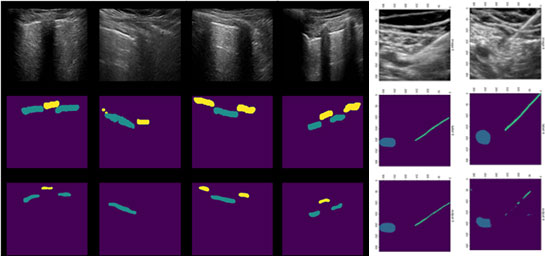
Once the relevant anatomy is identified, regions of interest are determined (e.g., the region encompassing the lung below the pleura for PTX cases). Our solution then analyzes those regions of interest to make a decision. For PTX, pixels from the ultrasound video are labeled as exhibiting respiratory motion (movement, labeled in blue) or not (labeled in green). Cases of no apparent respiratory movement are indicated as possible PTX cases.
The success of Phase I, which focused on detecting collapsed lungs, has led to a Phase II effort that will expand the application of AI to peripheral pain management, intubation, and traumatic brain injury. This collaboration exemplifies Kitware’s ability to drive innovation in medical AI, with our methods being adopted by other organizations involved in the program. We have decided to publicly release our source code and publish our Phase 1 work to help pave the way for future collaborations and advancements in the medical industry. We believe this DARPA POCUS project exemplifies how AI can revolutionize medical care on the battlefield and beyond.
If you’re interested in learning more about this project and Kitware’s capabilities in this area, please contact our team. You can also read our technical paper, which was published in The Journal of Trauma and Acute Care Surgery.
This material is based upon work supported by the Defense Advanced Research Projects Agency (DARPA) under Agreement No. HR00112190077.