Preserving Privacy, Advancing Research: Solutions for Medical Image Sharing

Collaboration and data sharing are crucial for advancing medical research and finding solutions to complex problems. However, it’s difficult to share medical data because it often contains protected health information (PHI), such as a patient’s name, birthday, or other personal identifiers. To protect patient privacy, the data needs to be de-identified before it can be shared.
Unfortunately, there are many complexities involved in anonymizing medical imaging data, particularly in the realm of whole slide imaging. Where most fields, such as radiology, have established standards for data anonymization, whole slide imaging lacks uniformity, posing significant challenges for redaction efforts.
One of the biggest problems is that not all of the metadata is irrelevant. For example, let’s say you ran two separate tests on the same patient that were a month apart. For whole slide imaging, this time span is a relevant data point that will help track the progression or remission of disease and should not be removed during the anonymization process.
Kitware’s de-identification software is capable of automatically detecting and redacting PHI while preserving essential clinical metadata within the whole slide images, no matter how large the file size. This software was developed as part of the National Institutes of Health’s ImageDePHI program, which focused on the Seer national pediatric cancer registry.
Given the rarity of pediatric cancers, it is much easier to identify patients based on their medical data. So Kitware saw an opportunity to bring our work on the WSI DeID project to a wider audience. Thus, ImageDePHI was born. During Phase 1, Kitware’s team focused on developing an automated redaction process that would protect patient privacy while retaining clinically relevant information. Our team built an algorithm that could reliably identify text and barcodes on whole slide images. We also developed a plugin tailored to Seer’s workflow that would allow this redaction technology to be more easily applied across other medical imaging fields.
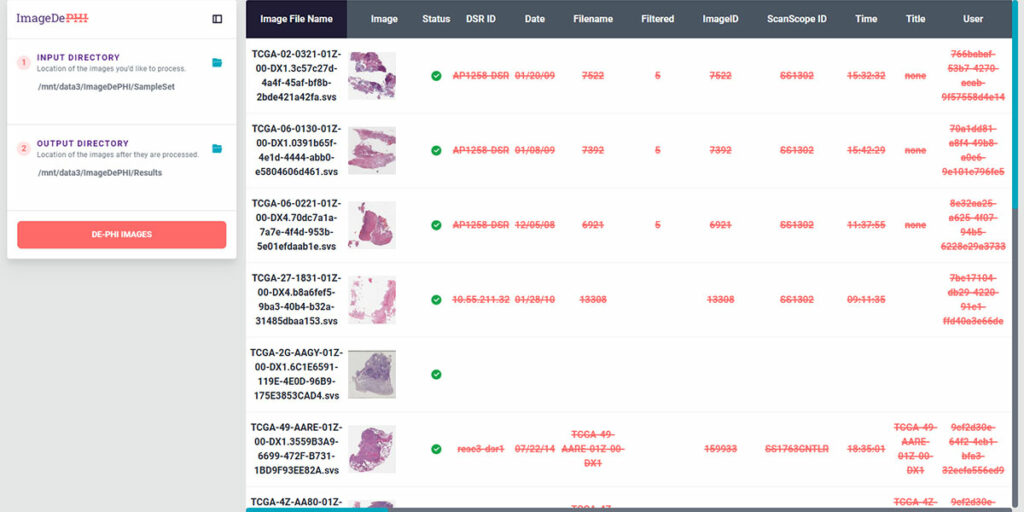
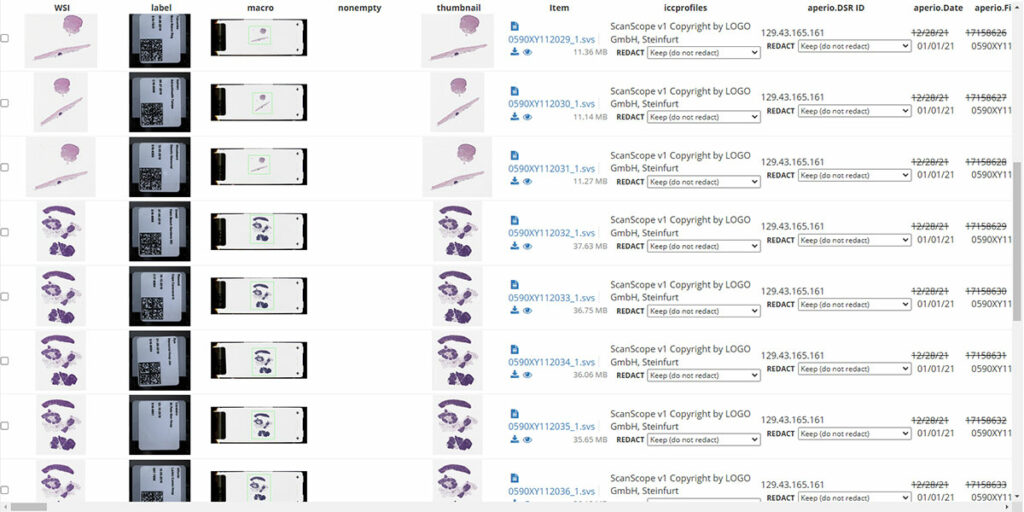
Kitware is continuing to develop this technology during Phase 2 of the program, including creating command-line interfaces and refining redaction algorithms based on user feedback. Once you have determined your redaction rules are correct, it can be simple to run the process on a directory of images. If an image has an unexpected piece of metadata the process will stop. We’ve also had to overcome unique challenges to ensure the technology could be used in real-world settings where the data isn’t always “perfect.” For example, a researcher may need to apply a new barcode on a slide over an existing one. However, the stickers may not line up with each other. We are refining our software so that it can still read, identify, and redact the information on both barcodes.
This work is crucial to empowering researchers to securely share and analyze medical imaging data. Kitware is passionate about advancing healthcare through open source technology. All of ImageDePHI’s cutting-edge tools and workflows are open source and available to any researcher that needs them. You can find everything you need in this Github Repo. There is a SEER-specific tool available in this repo. If you need assistance in using the software or would like to develop it further for your specific use case or product, please contact our team!