NimbusImage: an open-source image analysis platform for researchers in the life sciences
Imaging has led to major discoveries in the life sciences. With the advent of powerful imaging techniques and microscopes, we are now awash in data. However, the analysis of this heterogeneous data—at scale, with accuracy and reproducibility—has proven to be a major challenge.
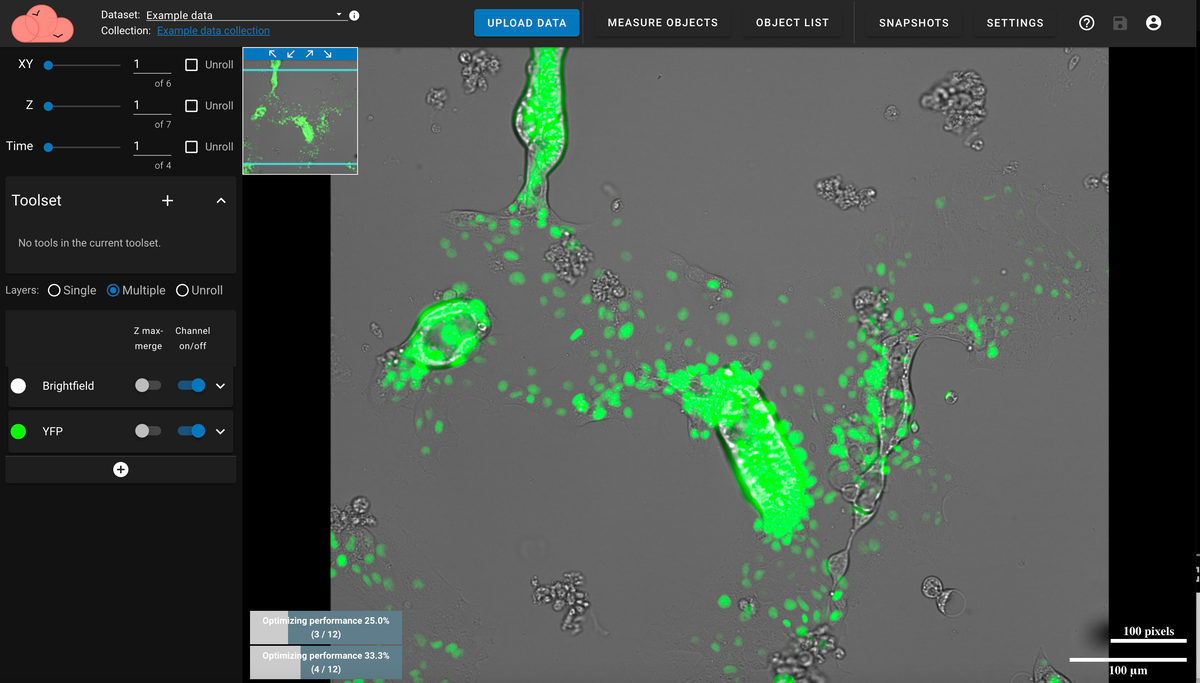
NimbusImage has been developed as a platform to empower scientists to analyze their image data and spend their time making discoveries rather than struggling to process their datasets.

NimbusImage is…
- Geared towards life sciences. The target audience is cell biologists who work with microscopy data. The focus has been fluorescence microscopy, but we are keen to expand to serve more communities over time.
- Web-based. NimbusImage lives in the cloud, requiring no installation. It can handle very large datasets without worrying about local storage and memory. NimbusImage enables fast and easy sharing of data and analyses.
- Scalable. NimbusImage was designed from the ground up to handle the viewing and analysis of large datasets. As microscopes have become capable of producing ever-increasing amounts of data, even just loading datasets can be a challenge. NimbusImage enables interaction with big datasets in the browser.
- Open source. NimbusImage is built in the open. Anybody can run their own image server. All the code is open for scrutiny.
- Fun. Image analysis is often a painful, mind-numbing task. We hope that NimbusImage will spark joy as you interact with your beautiful data.
What is special about NimbusImage
NimbusImage has a slightly different approach to image analysis than other tools. Most approaches to image analysis are built around pipelines, where you set a bunch of parameters, run a big dataset through the algorithms, and then end up with some numbers in a table. What is typically missing is the ability to visualize and interact with your analysis. For instance, maybe some region was out of focus and you want to exclude that. Or some spots were assigned to the wrong cell. Or some cell segmentation needs to be cut in half. NimbusImage allows you to use cutting-edge algorithms and interact directly with the output, making sure your analysis reflects your data accurately and injecting your knowledge and intuition back into your work.
Video 1: Uploading and navigating datasets
Video 2: Segment Anything Model (SAM) for quickly picking out objects
Watch more videos to learn how to find nuclei automatically or quantify fluorescence intensity.
Acknowledgments
NimbusImage has been developed by the lab of Arjun Raj at the University of Pennsylvania, a Professor in the Department of Bioengineering and the Department of Genetics in collaboration with Kitware, creators and maintainers of the open-source toolkits ITK and Girder.
And now…
Get started and try it out on nimbusimage.org!
Reach out to Arjun Raj if you want to start using NimbusImage in your lab.