Kitware applies machine learning technology to help pathologists efficiently analyze whole-slide images
Kitware produces software that uses AI and machine learning (AI/ML) technologies to advance the field of digital pathology in collaboration with academic researchers and clinicians. At the center of this effort is the Digital Slide Archive (DSA), an open-source platform, for centralized data management, sharing, annotating, and analyzing whole-slide imaging datasets, which continues to be actively developed and adopted by the digital pathology community. We report here a new technology, HistomicsStream, that the team has developed to make the process of working with whole-slide images (WSIs) more efficient. HistomicsStream is a python package that makes working with WSI data more approachable for computer vision and machine learning experts and will help significantly accelerate research in the field of computational pathology.
Understanding the Basics: What is histopathology?
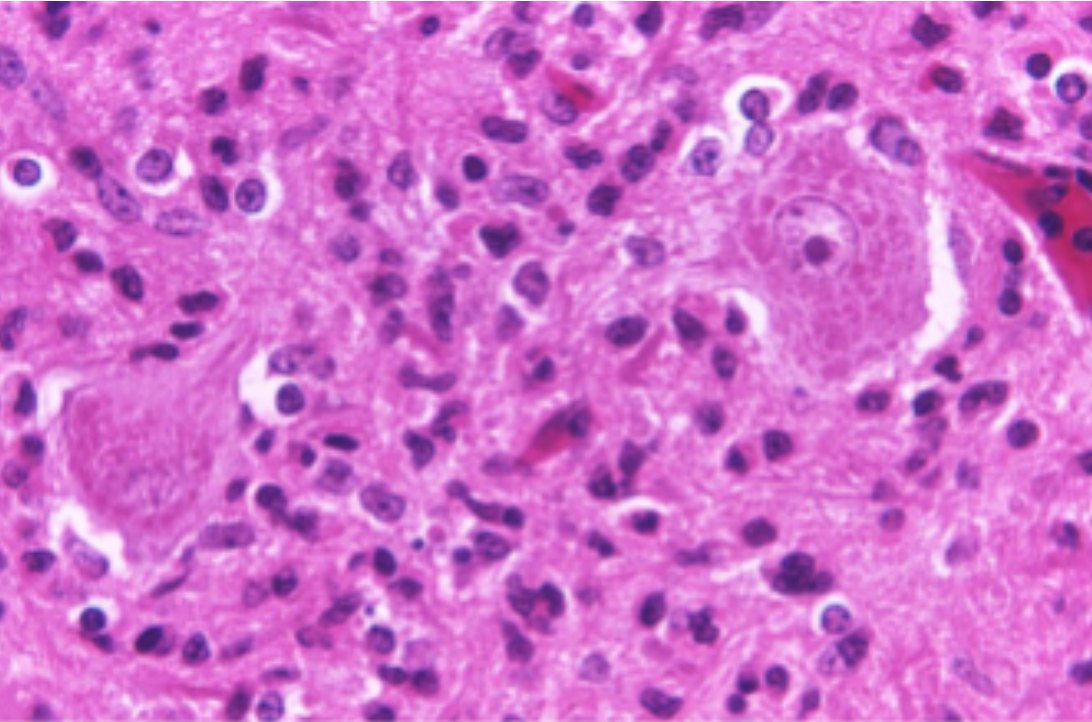
Histopathology is the study of biopsied tissues under the microscope for the purpose of diagnosing disease. Glass slides of tissue specimens are prepared by staining thin tissue slices with chemicals to highlight cellular structures for examination. Traditionally, pathologists have examined glass slides to look for telltale signs of disease, but WSIs that digitize the entire slide at high magnification are being used more regularly in diagnosis. Digitization also allows the utilization of computer vision algorithms to detect, classify, and measure structures in WSIs, helping to improve the quality and reproducibility of diagnosis.
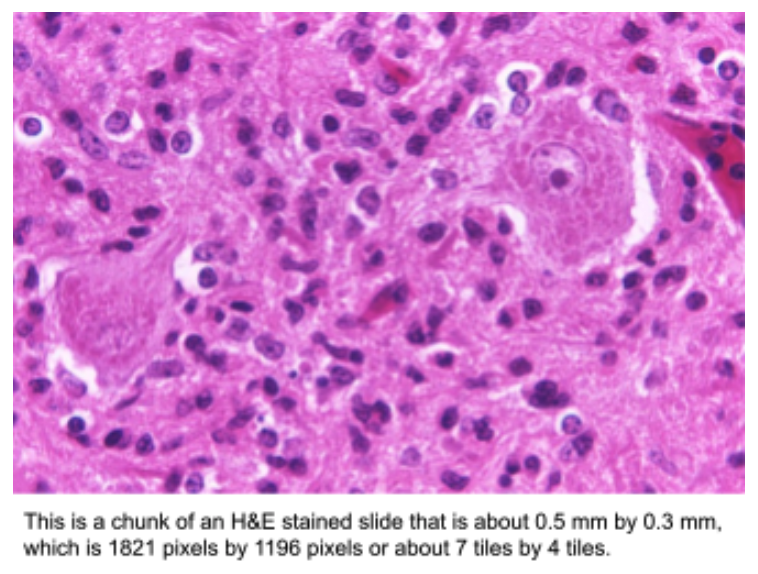
Applying AI/ML to Analyze WSI Data
Developing algorithms to analyze WSIs is challenging since popular machine learning frameworks and computing hardware are built for analyzing much smaller images. For example, a typical WSI with 120,000 x 80,000 pixels contains the equivalent of 191,000 non-overlapping tiles that are 224 x 224 pixels, which is a typical size used in machine learning frameworks. For machine learning purposes such as proposing regions of diagnostic value, these large images are usually broken up into these smaller tiles. Often, a group of tiles (e.g., 8×8 tiles) is read at once as a “chunk” from a datastore for efficiency purposes. However, especially with the prediction step of machine learning, simply reading these data from disk can be the most significant determinant of runtime performance.
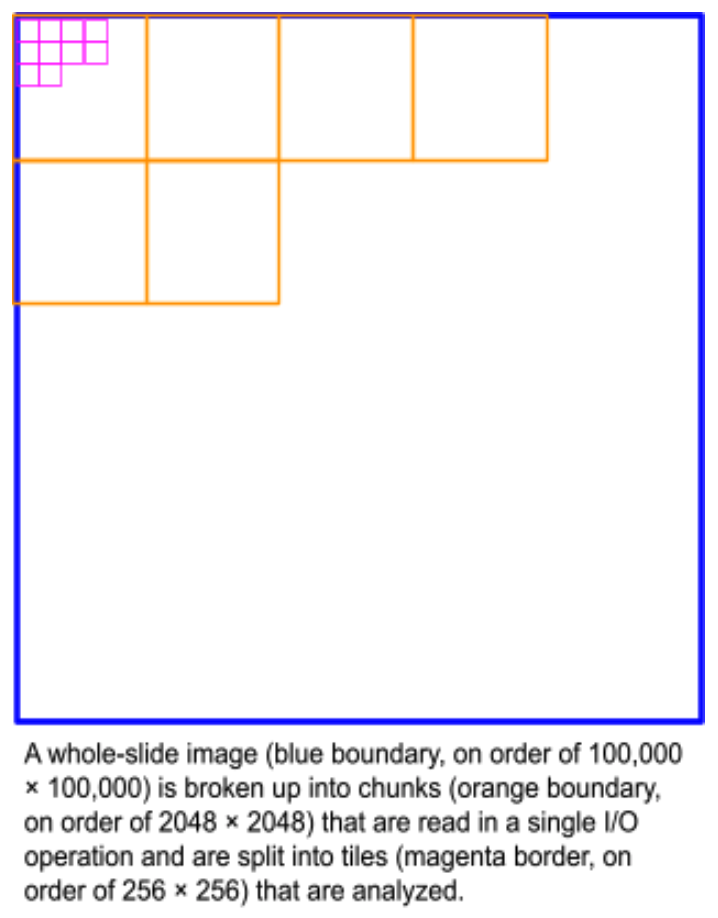
Several Python libraries, such as openslide and large-image, are capable of reading whole-slide images efficiently, but these are not readily compatible with machine-learning frameworks like TensorFlow. Additional power comes from packages such as Zarr, which distributes data from a single image across multiple files for parallel, cloud-native access, and writing. These packages are able to efficiently read a tile from anywhere within a whole-slide image without having to read the entire image. A common approach is to use these libraries to convert a WSI into many smaller files in PNG or JPG formats that can be read by the standard input routines in machine-learning frameworks. This approach has several important limitations when compared to directly reading data from WSIs. Firstly, changing the tile size requires re-tiling the image. Secondly, the data stored in WSIs is typically compressed using lossy compression like JPEG, and so transcoding or lossless compression should be used when generating the tiles. This can lead to a massive exploding in the amount of data used in a single experiment, which can number in the tens of thousands of WSIs. Our goal with HistomicsStream was to directly read from WSIs into machine-learning frameworks and to be able to leverage the prefetching and parallelization of their input routines.
About the Technology
Histomics_stream Python Package
histomics_stream is a Python package that enables efficient access to large datasets of whole-slide images for use in machine learning. Users specify the details of the data set, and the desired operating parameters, such as which images (and optionally a mask to specify which parts of them) will be processed, what size each tile should be, how much should tiles overlap, and how many tiles should be selected at random. histomics_stream will then create a TensorFlow execution graph that defines a TensorFlow Dataset. Together with TensorFlow’s scheduling and parallelism functionality, this execution graph simply and efficiently directs the reading of tiles from disk for efficient use in TensorFlow model operations. The TensorFlow Dataset created by histomics_stream is used directly in TensorFlow operations for machine learning, whether for model fitting, model evaluation, or use of a model to make predictions in novel input data.
The steps of histomics_stream are demonstrated in this Jupyter lab notebook (open in Google Colab) in the source distribution. The construction of a Python dictionary that describes the study data set is straightforward, and key steps are implemented by histomics_stream. Complexities from TensorFlow are seamlessly handled.
Improved Performance
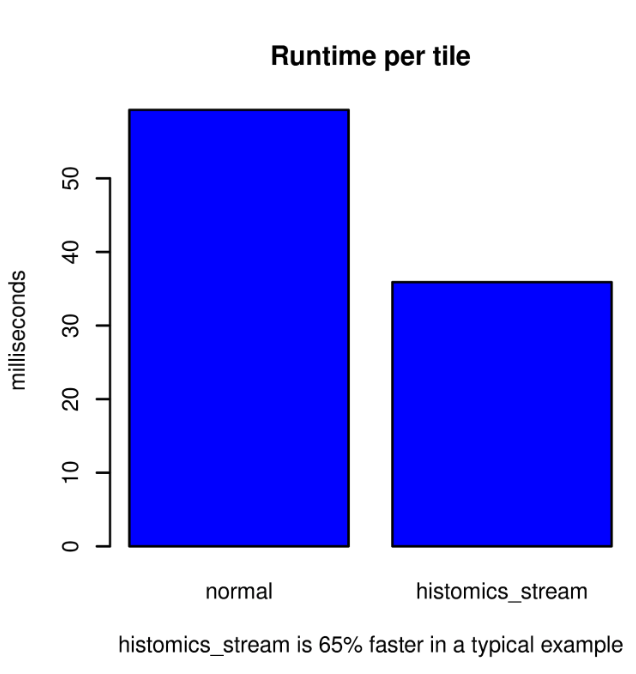
The histomics_stream package significantly improves runtime performance. In a typical example, reading a single whole-slide image that is 19,784 x 27,888 pixels as non-overlapping tiles that are 256 x 256 pixels produces a 77 x 108-pixel grid of 8,316 tiles. The large-image package is impressive in its ability to seamlessly read multiple file formats and efficiently read tiles from within large images. Using large-image only, the runtime is a quick 59.3 ms per tile, including reading and machine learning prediction, using a single GeForce RTX 2080 Ti. Using histomics_stream, this workflow runtime is reduced to 35.9 ms per tile, which is a 65% performance improvement. Much of the performance gain comes from reading data one chunk (2048 by 2048 pixels) at a time rather than one tile at a time. The additional performance gain comes from the reliance on TensorFlow for the scheduling of reads; TensorFlow’s graph execution schedules each read to optimize the overall performance of the workflow as a whole.
The TensorFlow graph execution interface can be challenging and unintuitive. Instead, bioinformatics model creators can use histomics_stream to specify the dataset that is to be analyzed. histomics_stream takes care of the TensorFlow execution graph creation and provides a significant runtime performance improvement. It is an efficient start to workflows such as those used for HistomicsTK and the DigitalSlideArchive.
Stay subscribed for follow-up posts, where we demonstrate how histomics_stream makes WSI breast cancer detection easy and performant, interfaces with cloud-native storage with Zarr, and leverages GPU-accelerated decoding with cuCIM.
Support from Kitware
Get expert help from the team behind histomics_stream, DSA, and many other informatics tools for data management, visualization, and analysis of digital pathology and medical image data. Kitware can support you throughout every stage of the process, from implementation to training. As a result of our long history of advancing the medical and biomedical fields, we are well-equipped to support your project and help you achieve your goals. Contact us for more information on how we can collaborate.
Acknowledgments
This work was funded by the National Institutes of Health National Cancer Institute Informatics Technologies for Cancer Research (NIH NCR ITCR) U01 grant 5U01CA220401-04 entitled “Informatics Tools for Quantitative Digital Pathology Profiling and Integrated Prognostic Modeling” with Dr. Lee A. D. Cooper (Northwestern University), Dr. Metin N. Gurcan (Wake Forest School of Medicine), and Dr. Christopher R. Flowers (Emory University) as principal investigators and Kitware as a subcontractor.