Introducing NIRFAST-Slicer 1.0
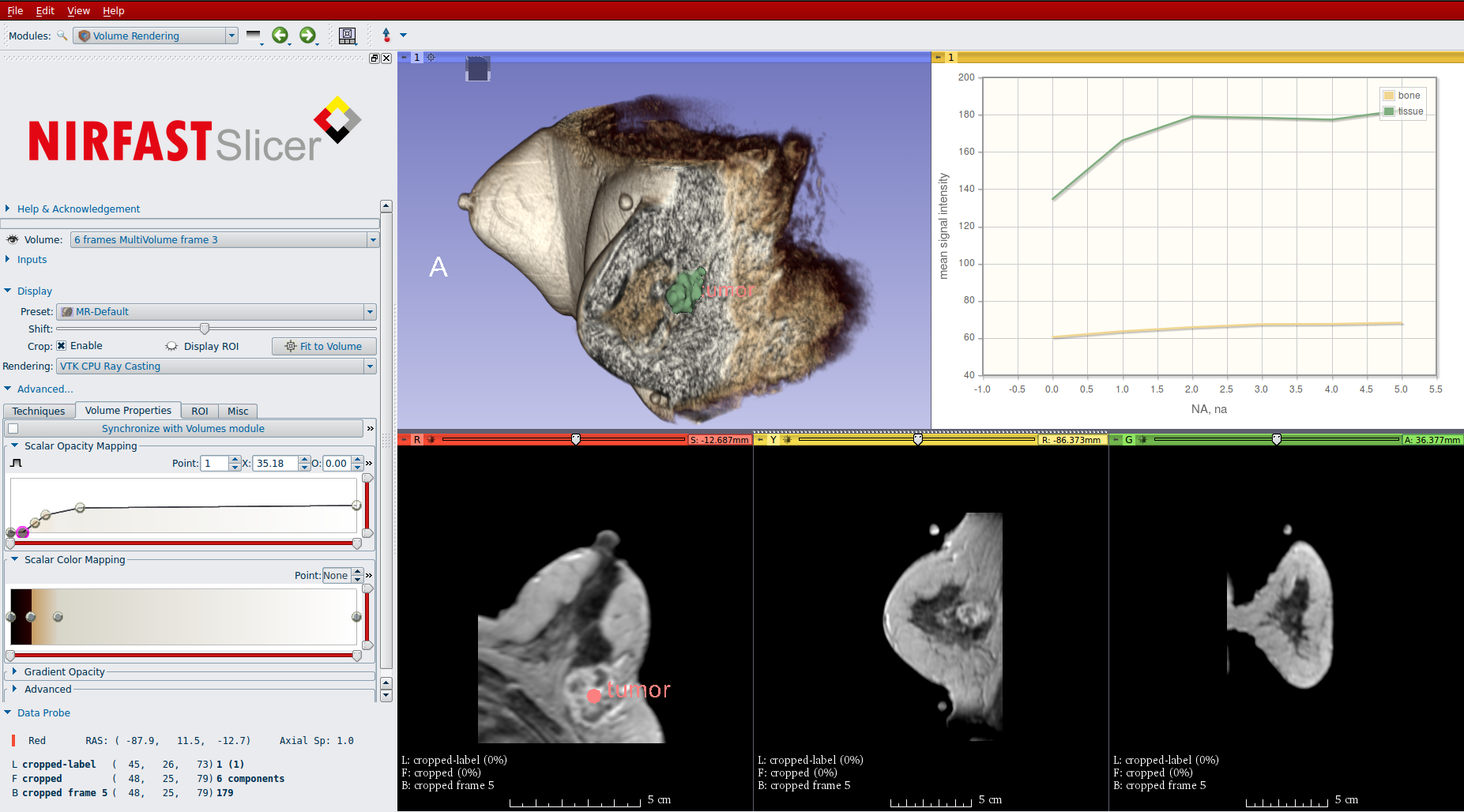
We are proud to unveil version 1.0 of the NIRFAST-Slicer application for optical imaging research. NIRFAST-Slicer brings together two powerful open-source toolkits used in the imaging research community: NIRFAST and 3D Slicer.
NIRFAST is an interactive open-source software application package that is widely used in the near-infrared imaging research community. It provides functionalities for modeling and reconstructing near-infrared light transport in tissue including standard single wavelength absorption and scatter, multi-wavelength spectrally constrained models and fluorescence models. It is used to model both light propagation in tissue (forward problem) and to recover internal optical parameters from optical data measured on the tissue surface (image reconstruction).
To improve optical parameter estimation, anatomical medical imaging modalities are used in NIRFAST. Accordingly, Kitware developed NIRFAST-Slicer in collaboration with Thayer School of Engineering at Dartmouth University and University of Birmingham. NIRFAST-Slicer provides powerful automated tissue segmentation tools that allow for the development of accurate finite element method (FEM) meshes. The application comes with a Matlab-bridge that facilitates data exchange between the 3D Slicer platform and the NIRFAST Matlab package. The segmentation results from NIRFAST-Slicer are used to set up boundary conditions and internal tissue structure for the FEM analysis in NIRFAST-Matlab.
A few custom Matlab modules are packaged in NIRFAST-Slicer 1.0. They are composed of an XML description file that defines the user interface through the Command Line Interface plugin mechanism and a Matlab file that calls NIRFAST-Matlab specific functions. This Matlab code is called through the MatlabCommander module, which first creates a connection to a Matlab server and then runs the execution of the code by specifying inputs set in the module interface inside NIRFAST-Slicer. The code also converts the outputs of the Matlab execution back in the 3D Slicer-based application. In addition, a Python module is packaged within NIRFAST-Slicer in order to read a mesh file created by NIRFAST-Matlab. It dynamically re-samples the internal optical parameters recovered after the reconstruction phase into 3D volumes in the original image space. Those can then be conveniently overlaid above the original image.
Installers for all platforms for NIRFAST-Slicer are available on http://bit.ly/NIRFASTSlicerPackages. For documentation and a step-by step tutorial on NIRFAST-Slicer, please see http://bit.ly/NIRFASTSlicerDocumentation. To go with the written documentation, check out this YouTube playlist, which shows each step of the NIRFAST-Slicer pipeline.
Kitware offers support for customizing and integrating its open-source software. To learn how to leverage solutions such as NIRFAST-Slicer and 3D Slicer, contact kitware(at)kitware(dot)com.
Acknowledgement:
Research reported in this publication was supported by NIH/NCI under Award Number R01CA184354. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

