How to Improve Software Quality in Open Source Projects: Part I – Overview of Continuous Integration (CI) and Software Process in PyNWB

Index To the Series
1. Overview of Continuous Integration (CI) and Software Process in PyNWB
2. Packaging PyNWB for Multiple Operating Systems and Python Versions
3. Code Coverage, Documentation and Style
Writing quality software requires you to have craftsmanship as well as technical expertise. While writing code is a demanding activity, you can automate portions of the day-to-day tasks to help reduce errors. By adding automated software quality processes, you can further increase the maintainability and readability of your project, which can help you build a community around and encourage contributions to open source software.
To support software quality, it is critical that you have a variety of tests check for errors at different levels of complexity in the development workflow. Other essential practices such as using small and atomic commits and writing intelligent commit messages will help you foster consistency and gain contributions to your project. To take your software to even higher levels of capability, performance and quality, you can practice continuous integration (CI).
CI automates critical software process tasks such as compiling software, running tests, releasing and deploying software and storing artifacts. In CI, these tasks are invoked persistently as you make changes to the software. CI builds may be triggered when you push a commit to a source code repository, for example, or when you merge a topic branch into your master branch.
In this blog series, we will explore the benefits that cloud-based CI provides. The actual examples are based on the software quality processes that we added to PyNWB, an open source software project. The examples present a guide for better understanding the tooling used in PyNWB.
PyNWB
PyNWB is a Python software library that provides a high-level application programming interface (API) for working with files stored in the NWB format. NWB is a unified data format for cellular-based neurophysiology data. It focuses on the dynamics of groups of neurons that are measured under a large range of experimental conditions. The NWB format comes from the Neurodata Without Borders: Neurophysiology (NWB:N) project, which consists of neuroscientists and software developers who recognize that the adoption of a unified data standard is an important step toward breaking down the barriers to sharing, reusing and reproducing data.
The goal is to have PyNWB be as accessible to as many researchers as possible. By adding automated tooling to the CI process, we enabled PyNWB to support Python 2 and Python 3; be usable on Windows, Mac and Linux operating systems; and be packaged to both PyPI and Conda-forge. To do so, we needed to test PyNWB on multiple platforms. Without automated tooling, it would have been far too much manual labor for us to keep up with our goal and pursue code development.
In addition to making PyNWB accessible, we aim to encourage good quality and consistency in the codebase to ensure that PyNWB is maintainable. As a result, we added style enforcement in our CI tests, and we automatically build the documentation as part of our CI process.
Tool Summary
As we progress through this blog series, we will guide you to add tools to a new or existing software project. To do so, we will reference a large number of technologies, which include the following:
- CircleCI, a cloud CI platform that PyNWB uses to run Linux tests;
- AppVeyor, a cloud CI provider that PyNWB uses to run Windows tests;
- Travis CI, a cloud CI service that PyNWB uses to run Mac OS tests;
- versioneer, a tool for adding semantic versioning;
- tox, an automated test runner for different Python versions;
- PEP8 Speaks, a Github application for checking pep8 standards on pull requests;
- Sphinx, a tool for generating documentation;
- Requires.io, a tool for checking the status of requirements;
- Six, a Python package for creating compatibility between Python2 and Python3;
- Flake8, a Python tool for style enforcement;
- Codecov, a tool for reporting code coverage;
- PyPI, a Python Package Index, a repository of Python packages used by pip;
- Conda-forge, a Python package index used by Conda; and
- Read the Docs, hosting documentation generated by Sphinx.
Many of these cloud-based CI services will create badges for the state of your software project, which you can use to display the current status in a top-level view. For PyNWB, these badges are defined in the project’s README file. They result in a page that is rendered when you access the GitHub project.
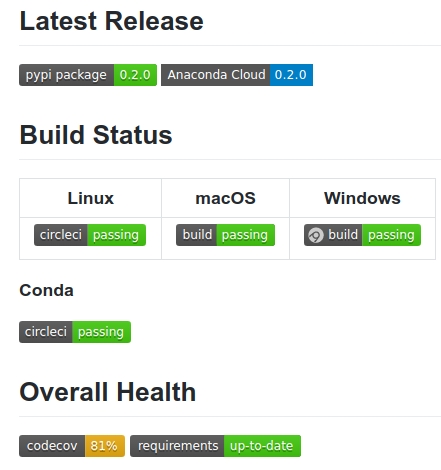
The page also includes a download button, a list of commits and an overview. Additional documentation on the project is available at https://pynwb.readthedocs.io/en/latest.
Future Posts
Moving forward, this blog series will offer a general overview of the PyNWB CI process, but it will not cover all of the setup steps in detail. The creation and management of user accounts, as examples, may be more idiosyncratic based on the needs of specific projects. Instead, we will focus on these topics:
- cross-operating system support, Python 2 and Python 3 support, source distributions and dependency versioning;
- code coverage, documentation and style; and
- real-world considerations, including the challenges of maintaining services.
To be clear, simply using the techniques described in this series won’t result in good code. Writing maintainable and understandable code is like walking on a tightrope, and the tools in this post are like an umbrella. They can help you reach your destination by improving your balance, but they are not enough to save you from falling.
Acknowledgements
Many people contributed to the software quality process of PyNWB. Thanks to Andrew Tritt for creating the Python test suite; to Andrew Tritt and Oliver Ruebel from Lawrence Berkeley National Laboratory for core development of PyNWB and for setting up the accounts, permissions and discussions; to Chris Martin for coordinating resources between The Kavli Foundation and NWB; to Nick Cain and Justin Kiggins from the Allen Institute for Brain Science for planning and porting PyNWB to Python 2 using the six library; and to the entire NWB team for discussions and collaborative development.