Digital Slide Archive, Large Image, and HistomicsTK: Open-source Informatics Tools for Management, Visualization, and Analysis of Digital Histopathology Data
This blog article presents an overview of the work being done in collaboration with Emory University on an ongoing NIH grant funded project for the development of informatics tools for the web-based management, visualization, and analytics of digital histopathology data. This project is being executed by a tight collaboration between the members of the Medical Computing and Data Analytics teams at Kitware and the research labs of our collaborators from Emory University.
Introduction
Histopathology refers to the study of the presence, extent, and progression of a disease through microscopic examination of thin sections of biopsied tissue that are chemically processed and fixed onto glass slides and dyed with one or more stains to highlight different cellular/tissue components (e.g. cell nuclei, cell membrane, cytoplasm) and antigens/proteins (e.g. Ki-67 indicating cell proliferation) of interest. It is regarded as the gold standard in clinical diagnosis, grading, and prognosis of several diseases including most types of cancer.
Source: NY Times
Pathologists visually scout across the tissue section under a microscope and examine the regularities in the appearance and architecture of a variety of biological structures (e.g. cells, glands) to determine if it is malignant and assess the level of malignancy. This process is very time consuming and its success depends heavily on the expertise and experience of the pathologist with substantial intra-observer and inter-observer variation. The recent emergence and increased adoption of cost and time efficient whole-slide image (WSI) scanners that can capture digital images of an entire tissue section at a cellular resolution within a few minutes has opened the door for the development of computer-aided image analysis algorithms to improve the efficiency, objectivity, reproducibility and accuracy of the diagnostic/prognostic process.
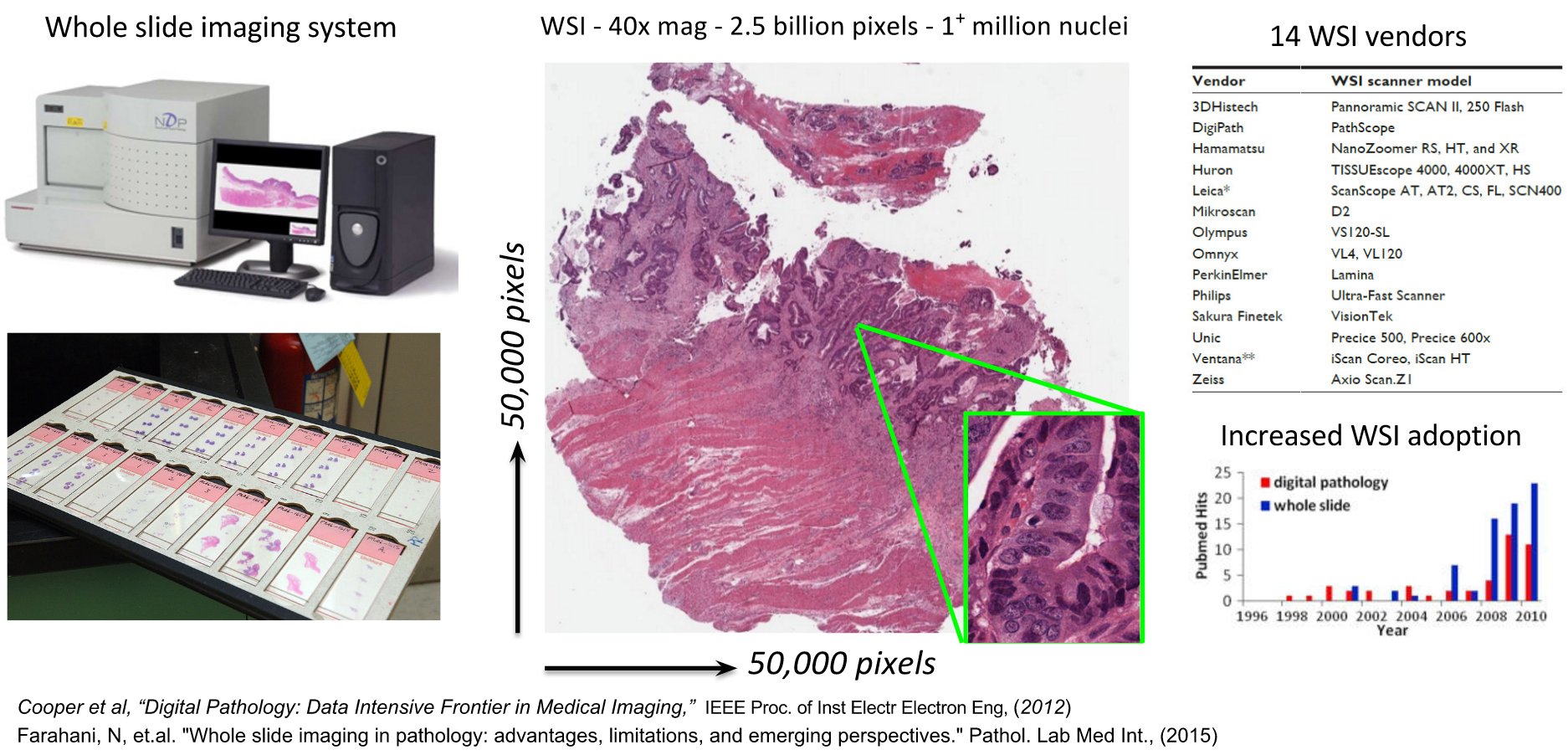
Whole slide images often contain over a billion RGB pixels and cancer studies may generate hundreds or thousands of such images. Due to their large size, these images are stored in a pyramidal structure (image is stored at increasing magnification at each successive level of the pyramid) for faster visual (pan-zoom) navigation and to facilitate multi-scale analysis. Enabling the management, visualization and analysis of whole-slide images requires considerable software infrastructure and presents significant challenges for software engineering. While multiple commercial tools are being actively developed to serve many of these needs, they come at a considerable cost, are closed-source and cannot be extended or modified to address specific user needs.
With funding from a U24 grant (U24-CA194362-01) from the Information Technology for Cancer Research program of the National Cancer Institute (NCI) and in collaboration with the research labs of the Principal Investigators (PI) Dr. David Gutman and Dr. Lee Cooper from Emory university, we are actively developing a suite of open-source web-based informatics tools to serve the data management, visualization, and analysis needs in the domain of of the massive and rapidly growing collections of data in the domain of digital pathology. Below is a brief description of each of these tools.
Our solutions in the making:
- Digital Slide Archive (DSA) for data management: DSA is a web-based platform for the aggregation, management, and dissemination of digital histopathology images and associated clinical and genomic metadata. DSA is being developed as a successor to the Cancer Digital Slide Archive (CDSA) – a user-friendly web-based tool developed by our collaborators at Emory university that allows users to browse, search and visualize the digital pathology data from a large public data resource called The Cancer Genome Atlas (TCGA) that contains over 24,000 whole slide images spanning 33 types of cancer from over 11,000 patients along with associated pathology reports, clinical data, treatment and genomic information. DSA builds upon on an open-source web-based data management platform, actively developed by Kitware, called Girder that provides essential general-purpose data management capabilities such as uniform RESTful access to data located on a variety of back-end storage engines (e.g. Amazon S3, GridFS, HDFS, Native file system); user/group authentication and management; fine-grained access control of data; and a well-designed plugin development framework for building web-based data analytics applications. In addition to inheriting these, DSA intends to provide many additional features specific to the digital pathology domain, such as a thumbnail gallery views of whole-slide images in a collection, an embedded multi-resolution image viewer for visualizing whole-slide images overlaid with annotations, association of images with custom metadata, and a faceted search interface that enables searching/filtering of data collections by metadata and creation of cohorts. Below is a demo video showcasing the features currently available in DSA.
- Large image for visualization: Large image is a Girder plugin for the visualization of large multi-resolution (whole-slide) images overlaid with annotations using multiple back end viewers (e.g. GeoJS, SlideAtlas, OpenSeaDragon, OpenLayers, Leaflet) with pan-zoom capabilities. It also functions as a standalone Python toolkit (built on top of OpenSlide) for reading/writing large multi-resolution (whole-slide) images at different scales in a tiled fashion. It handles many WSI file formats and provides conversion methods for formats that it doesn’t directly handle or for images that aren’t already stored as multiple resolutions for efficient access. It automatically extracts some metadata from the WSI images such as magnification and associated label and macro images. Lastly, it also includes a JSON schema for various kinds of annotations (e.g. points, lines, rectangles, ellipses, polygons, heat-maps, image-overlays) and will provide a Python API to generate and read these annotations.
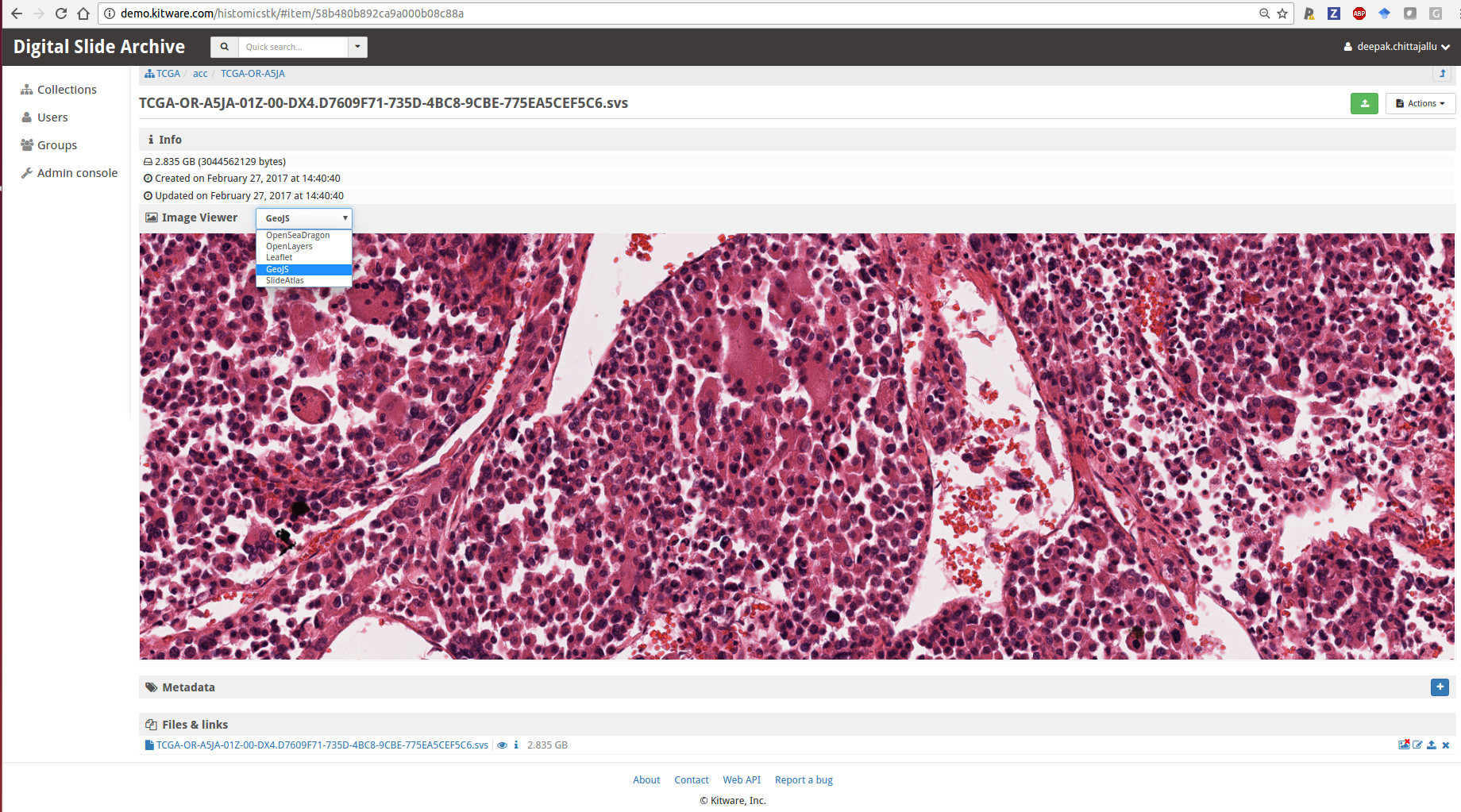
A multi-resolution viewer embedded within DSA for visualizing whole-slide images using one of several viewers - HistomicsTK for data analytics: HistomicsTK intends to serve both as a standalone python toolkit and as a web-based platform, developed as a server-side Girder plugin, for the analysis of whole-slide histopathology images in association with clinical and genomic data.
- As a standalone Python toolkit for supporting algorithm research and development: In this mode, HistomicsTK intends to provide a toolkit with implementations of state-of-the-art algorithms for common domain specific image analysis tasks such as: color/stain normalization, stain separation aka color deconvolution, cell/nuclei/membrane/tissue segmentation, extraction of image-based features at the object and image-level, cell/nuclei/tissue/image classification, cancer grading, and survival prediction.
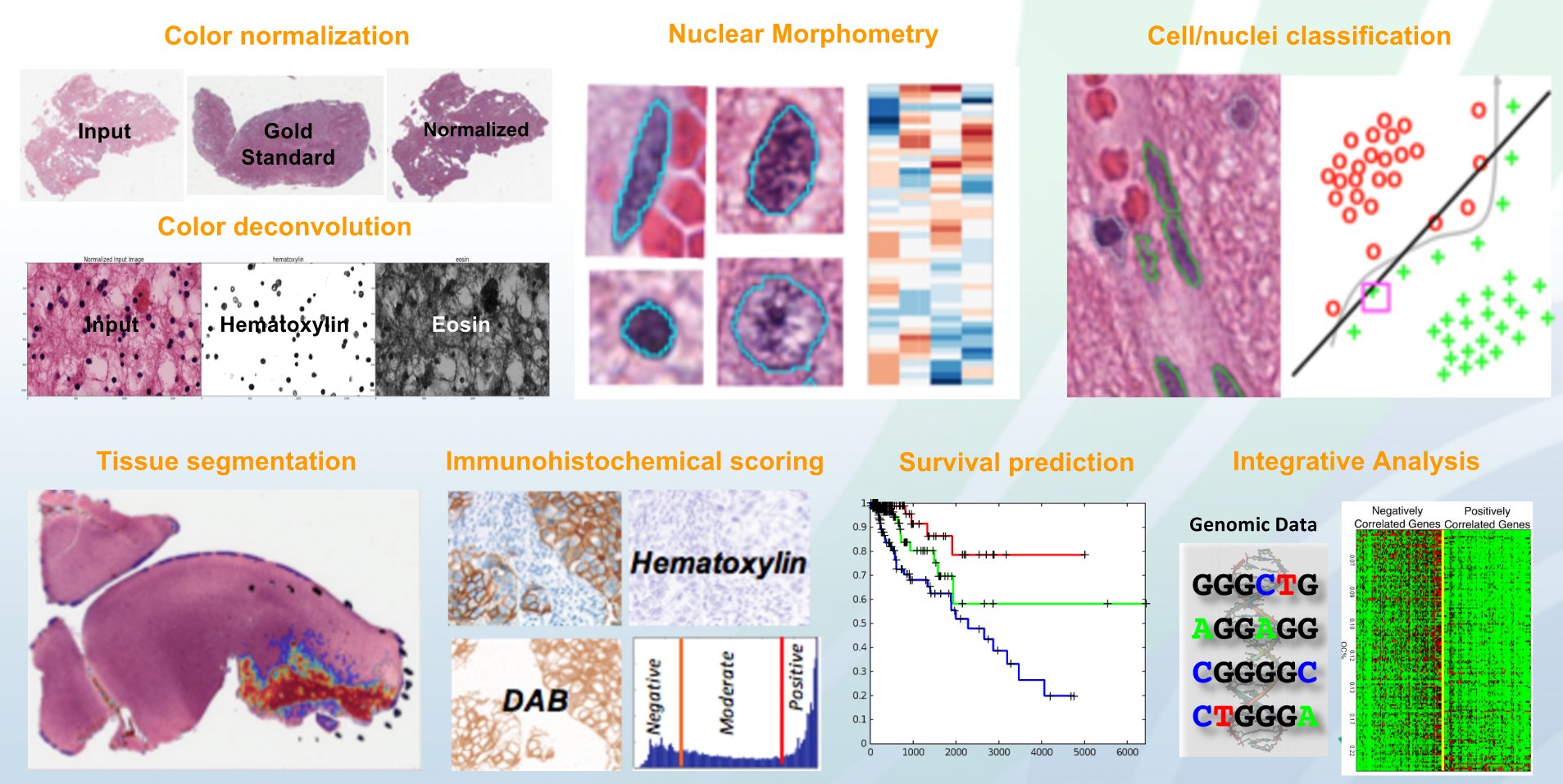
HistomicsTK – Algorithms - As a server-side Girder plugin for facilitating web-based analysis: In this mode, HistomicsTK intends to provide a collaborative platform that enables: a) Algorithm research labs to easily disseminate their analytics solutions over the web in a portable fashion for wider use by the community and b) Pathologists/biologists to easily test/apply state-of-the-art analytics solutions on their data over the web. By being developed to function as a Girder plugin, it can seamlessly inter-operate with and leverage the functionality of any other Girder plugin including DSA. It currently provides the following features:
- Automatic generation of REST endpoints and a user-friendly web UI for the execution of command-line analysis modules (containerized with Docker for portability) over the web using a Girder plugin called Slicer-cli-web.
- Distributed execution and monitoring of analysis tasks using a Girder plugin called Girder-worker and a Python toolkit called Dask.
- Multi-resolution visualization of whole-slide histopathology images overlaid with human/algorithm generated annotations using a Girder plugin called Large-image.
- A custom web-UI to interactively draw annotations on the images.
- A web-based analytics dashboard for interactively running analysis modules on data within DSA and visualizing the results.
In the future, we plan to develop a well-defined mechanism to chain a series of atomic analytics modules into full-fledged end-to-end pipelines, and distributed execution of analytics pipelines on a large batch of datasets. We are also planning to develop and integrate a web-based Active Learning system for semi-supervised training of machine learning models for a variety of predictive tasks. A demonstration of a preliminary prototype of this system developed in the lab of our collaborator Dr. Lee Cooper from Emory University can be seen in this video screencast.

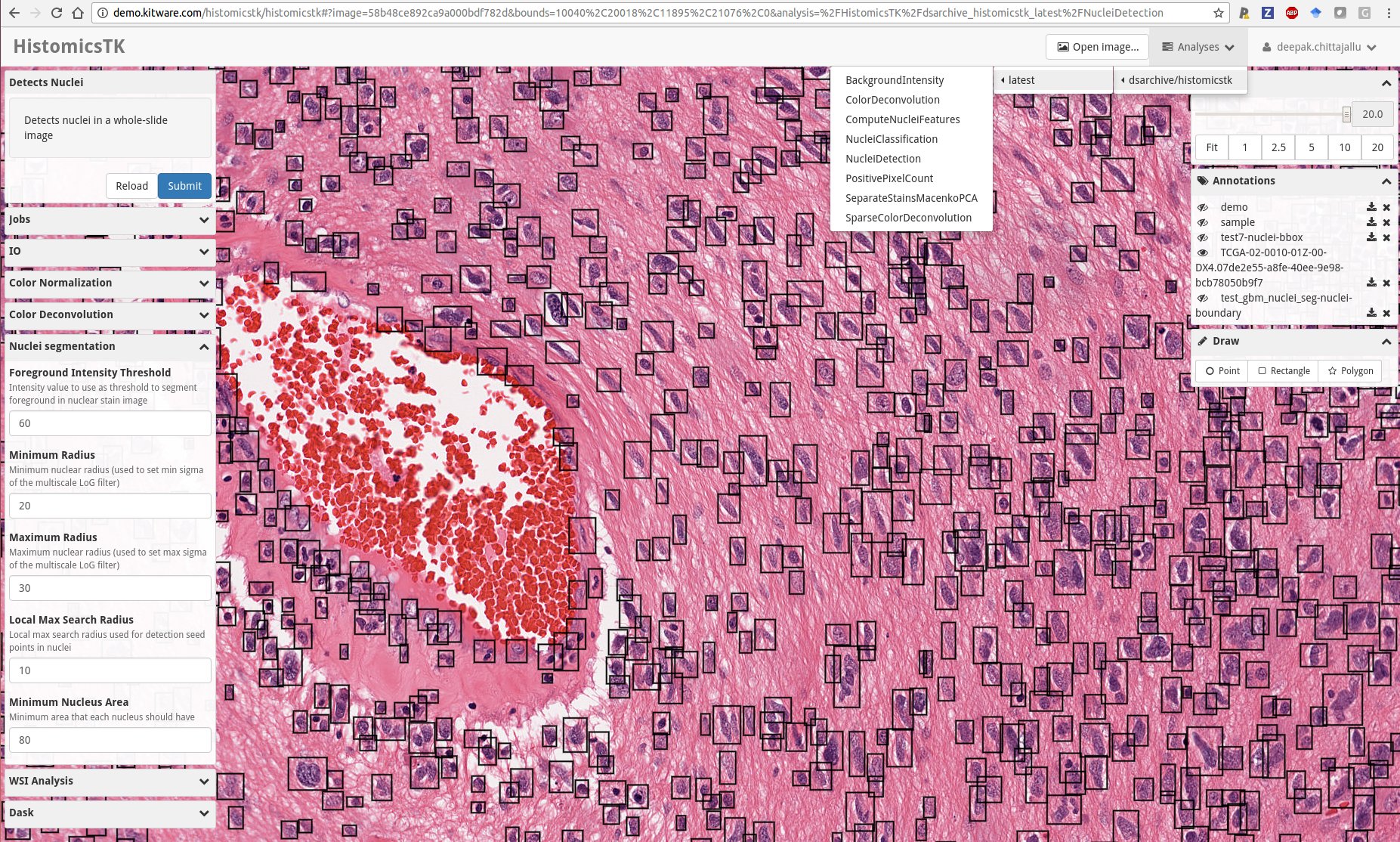
Snapshot of the web-based data analytics dashboard of HistomicsTK showcasing several features: (i) Drop-down menu showing Slicer CLIs in HistomicsTK’s docker image, (ii) left-panel showing the web UI autogenerated using slicer-cli-web to set parameters and run one of the CLIs, (iii) An embedded multi-resolution viewer provided by large-image for visualizing whole-slide images overlaid with annotations, (iv) bottom-right panel for interactively drawing various kinds of annotations on the images.
Conclusion
We use vagrant and docker to simplify the installation (see instructions here) of all the aforementioned informatics tools and their dependencies. A demo instance of these tools is hosted here for which a guest account can be requested. For those who seek to leverage any of these informatics tools, Kitware offers consulting and support services to help tailor our solutions to address user specific needs, create custom software, and/or conduct R&D. To learn more about the services offered by Kitware please contact us at kitware@kitware.com.Acknowledgement
This work is funded by the NCI’s ITCR U24 grant U24-CA194362-01 entitled “Advanced Development of an Open-source Platform for Web-based Integrative Digital Image Analysis in Cancer” with Dr. David Gutman and Dr. Lee Cooper from Emory University as PIs and Kitware as a subcontractor. - As a standalone Python toolkit for supporting algorithm research and development: In this mode, HistomicsTK intends to provide a toolkit with implementations of state-of-the-art algorithms for common domain specific image analysis tasks such as: color/stain normalization, stain separation aka color deconvolution, cell/nuclei/membrane/tissue segmentation, extraction of image-based features at the object and image-level, cell/nuclei/tissue/image classification, cancer grading, and survival prediction.
Is the MICCAI BRATS 2012: Multimodal Brain Tumor Segmentation Challenge dataset still available for download?