Creating Python Packages for Slicer Extensions: Lessons from the Cell Locator Project
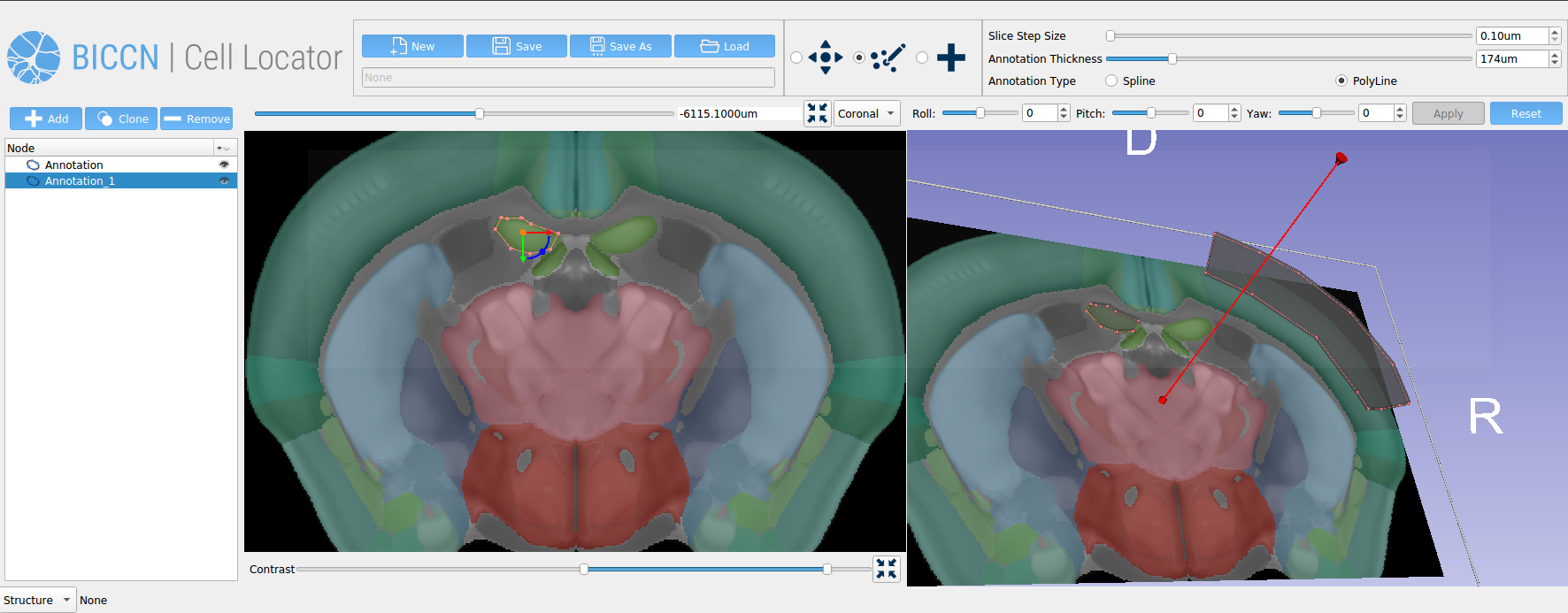
In our previous article, we described how our team at Kitware developed Cell Locator, an annotation tool that uses 3D Slicer to locate brain samples in 3D reference atlases. Cell Locator integrates with the workflow of the Allen Institute for Brain Science and supports neuroscience research by enabling annotations of experimental results.
While Cell Locator is built on the rich feature set of Slicer, it comes with a cost in startup time, memory requirements, and portability. To address these challenges, we developed cell-locator-cli, a Python package that is part of the Cell Locator tool suite and can be installed independently of Cell Locator or Slicer. This CLI tool enables certain batch-processing features and can be run from the command-line interface of a variety of operating systems. In this blog post, we detail some of our organizational decisions, such as the process of keeping the CLI code located in the same repository as Cell Locator, and the build process we used to publish this package.
Cell Locator
Cell Locator is an annotation tool used to more easily locate brain samples in the 3D reference atlases that we have developed for our partners at the Allen Institute for Brain Science and BRAIN Initiative Cell Census Network (BICCN) (Figure 1). It is an open source, cross-platform desktop application based on 3D Slicer, which integrates with the Allen Institute’s workflow and web services to support neuroscience research. Here, we present our software development and collaboration strategies.
One difficulty with this approach is that Cell Locator deals with a file format that is modified from native Slicer Markups, to include additional metadata and shape information for these annotations. Also note that neither this modified format, nor the native Slicer format, are supported natively by VTK or other external processors. This leads to a need for external tools to consume this file format so that processing can be done in batch without the overhead of a running Slicer instance.
To support this use case, we created a Python package cell-locator-cli that contains several CLI tools and a Python library for loading and processing these annotation files from a pure-Python context. Because this package is so tightly coupled with the Cell Locator tool, we keep the source code for both in the same repository, which leads to some new challenges in adopting packaging tools where this is not the default configuration.
Python Packaging
For new packages, we prefer using pyproject.toml to specify package metadata such as dependencies, entrypoints, acknowledgements, and links. Recent versions of pip and setuptools have native support for pyproject-based packages; for example new versions of setuptools support editable installations with pyproject-based packages via pip install -e . for easier development and debugging. setuptools has comprehensive documentation on the supported features of the pyproject.toml format.
In a standard Python project, the pyproject.toml metadata file is usually located at the repository root. However, since we want to keep our Python package alongside our Slicer extension, maintaining the metadata file at the root becomes inconvenient. To solve this, we create a subdirectory to use as the package root, and place our pyproject.toml there instead.
To publish our package, we use the official PyPI tool: twine. Since we host the project repository on GitHub, we also use GitHub Actions for automated tests and deployment. We use the python-publish workflow, again considering the cli-v tag prefix.
Versioning
By placing our Python package in a subdirectory of our Slicer extension, versioning is a bit more complex. We need a way to indicate a different version number for our Python package than for our Slicer extension that doesn’t confuse our build tools and automation. The solution here is to use a prefix in our version tags; in our case, we use the prefix cli-v. In this way, our build tools differentiate between Cell Locator version v0.3.0 and Python package version cli-v0.1.0.
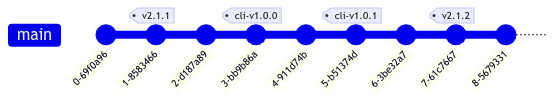
Since we use setuptools as our build backend, we also use setuptools-scm to automatically identify package versions based on git commit history. So in the above example, the current version of Cell Locator should be 2.1.2, while the published version of the companion package on PyPI should be 1.0.1. Critically, we don’t want setuptools-scm to incorrectly infer version 2.1.2 for the companion package. We enforce this with a custom git root and describe command, defined in pyproject.toml:
[tool.setuptools_scm]
root = '..'
git_describe_command = [
'git', 'describe', '--dirty', '--tags', '--long', '--match', 'cli-v*'
]This config leads setuptools-scm to only consider those tags with cli-v prefix, resulting in correct versioning. The version of Cell Locator itself is explicitly set, but a similar solution would be required in CMake projects with similar version number inference.
Lastly, since the version parsing logic implemented in setuptools-scm looks for a string of the form vX.Y.Z, this ensures the companion package is effectively released as X.Y.Z.
Documentation
Cell Locator documentation is hosted on ReadTheDocs. As before, the CLI is tightly coupled to the core application. To avoid fragmenting the documentation we include the cell-locator-cli documentation in a subsection of the Cell Locator page. We prefer to combine the Sphinx documentation generator with the myst_parser Sphinx extension to enable using markdown as our markup language. In that particular instance, we used sphinx-rtd-theme; however, for new projects we recommend using the furo theme combined with the sphinx-design extension.
Since we use the same docs page, it is not necessary to set a tag prefix in the ReadTheDocs config; all the documentation must be rebuilt for every change. However, ReadTheDocs does support tag prefixes for those cases where documentation should only be rebuilt for certain version changes. Note that tags are supported in the web config interface, but not in the new .readthedocs.yml file. See configuration file details for more information.
Acknowledgements
Research reported in this publication was supported by the National Institute Of Mental Health of the National Institutes of Health under Award Numbers U01MH114812 (PI. E. Lein), U19MH114830 (PI. H. Zeng), and U24MH114827 (PIs M. Hawrylycz, L. Ng). Additionally, development of the software process infrastructure was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under grant number R24 GM136986 (PIs C. R. Johnson, R. S. MacLeod, R. T. Whitaker). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Research Resource Identifiers (#RRID) referenced in this publication are BICCN Cell Locator (BICCN Cell Locator, RRID:SCR_019264), BRAIN Initiative Cell Census Network (BICCN, RRID:SCR_015820), Allen Mouse Common Coordinate Framework (CCF) (Allen Mouse Brain Common Coordinate Framework, RRID:SCR_020999) and Allen Human Reference Atlas (Allen Human Reference Atlas, 3D, 2020, RRID:SCR_017764).
We would like to thank the following past and present members of the Allen Institute for Brain Science team for their contributions to this project from administration and planning, to providing early feedback and improvement suggestions: David Feng, Stephanie Mok, Nick Dee, Tamara Caper, Rachel Dalley, Arielle Leon, Rusty Mann, Zach Madigan, Rob Young, Josh Royall, Scott Daniels, Katherine Baker, Hongkui Zeng, Song-Lin Ding and Ed Lein.