3D Slicer’s Liver Segmentation: A Look at the New Features
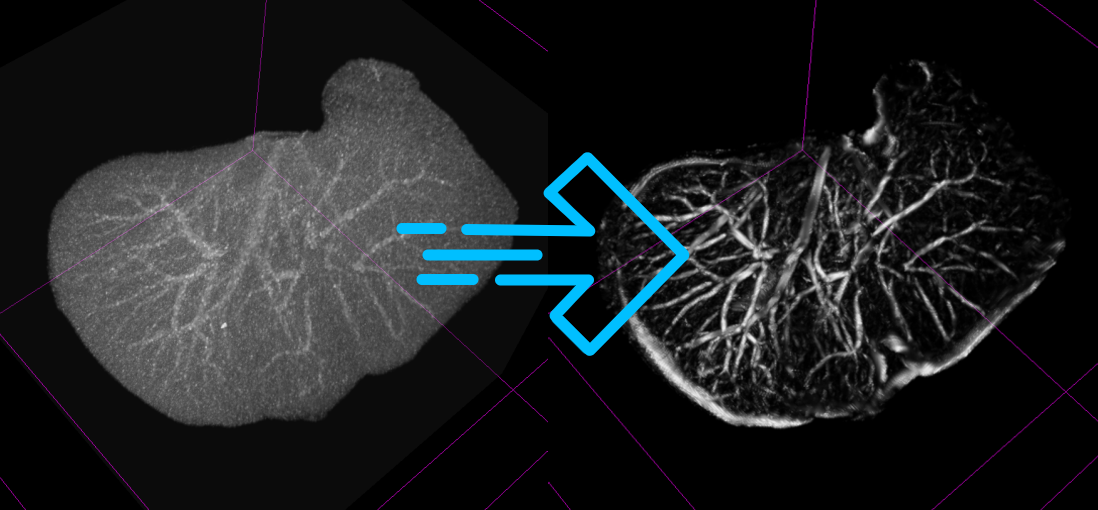
3D Slicer, the open-source software platform for medical image informatics, image processing, and three-dimensional visualization, continues to evolve with the addition of new extensions that enhance its capabilities in liver anatomy segmentation. Building upon the foundation laid by the SlicerRVXLiverSegmentation extension, the recent updates have introduced significant improvements and new functionalities that promise to streamline the workflow for medical professionals and researchers.
SlicerRVXLiverSegmentation v1.1.0: What’s New?
The SlicerRVXLiverSegmentation extension, initially designed to facilitate the annotation of liver anatomy from DICOM data, has received a noteworthy update with its v1.1.0 release. This version introduces enhancements that further aid in the segmentation of liver, liver vessels, and liver tumors, making the process more efficient and user-friendly.
Aside from various fixes, the new version includes an updated Liver segmentation module which now handles injected T1 MRI images in addition to CT images. This is available in the segment editor options using the modality drop down menu.
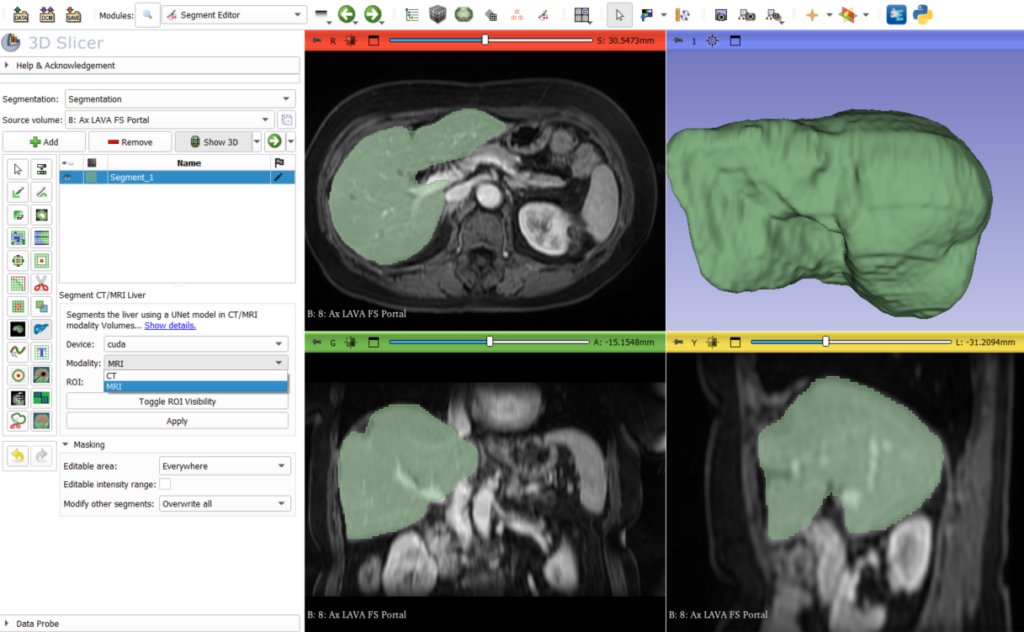
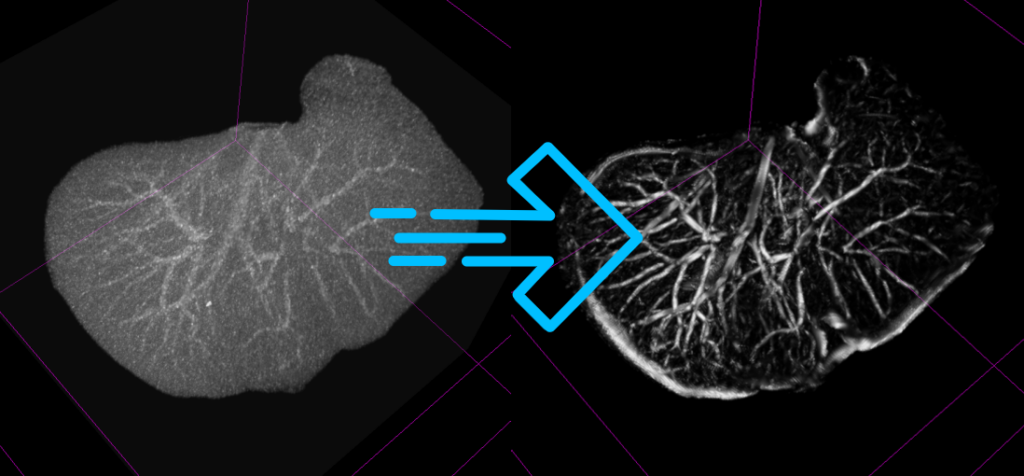
Introducing SlicerRVXVesselnessFilters
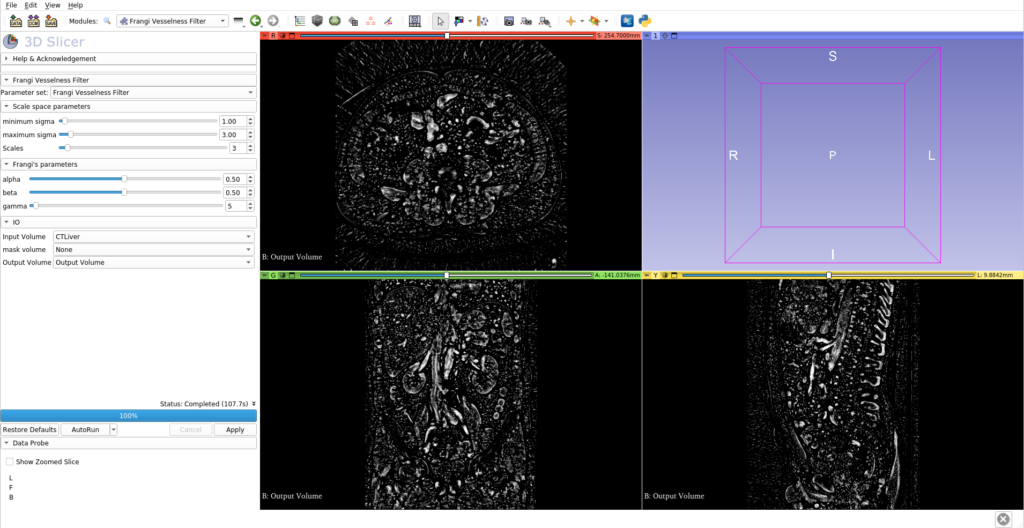
In addition to the updates to the liver segmentation extension, the 3D Slicer community has welcomed the SlicerRVXVesselnessFilters extension. This new addition brings a suite of vesselness filters tested specifically for liver vessel segmentation, offering a range of options to suit different imaging scenarios and requirements.
Highlights of the Vesselness Filters:
- Multiple Filter Options: Users can choose from several Hessian-based and morphological vesselness filters, each with its own strengths and applications.
- Command Line Interface (CLI) Extensions: The filters are accessible via CLI, making them easy to integrate into automated workflows and batch processing.
- Benchmarked Performance: The filters have undergone rigorous testing to ensure their effectiveness in various liver vessel segmentation tasks.
Conclusion
The continuous development of 3D Slicer’s extensions, such as SlicerRVXLiverSegmentation and SlicerRVXVesselnessFilters, demonstrates the commitment of the medical imaging community to improving the tools available for research and clinical practice. These updates not only enhance the functionality of 3D Slicer but also contribute to the advancement of medical research and patient care.
You can download those extesions from Slicer’s extension manager:
Reach us directly at kitware@kitware.fr if you need further information.
Credits
Kitware’s partners on this project are the LIRIS laboratory, Pascal institut, CReSTIC laboratory, and the CHU Clermont Ferrand.
This work was funded by the French Agence Nationale de la Recherche (grant ANR-18-CE45-0018, project R-Vessel-X, https://drv.uca.fr/ingenieriebr-de-projets/projets-finances/projets-termines/r-vessel-x). Antoine Vacavant is coordinator of the project (CHU Clermont Ferrand, Pascal institut).
I’m really excited to encounter such an excellent liver segmentation module while attempting liver segmentation – it’s awesome! It achieves perfect segmentation in just 3 seconds. However, I’m facing an issue: When using the SlicerRVXLiverSegmentation v1.1.0 extension, the 3D Viewer displays both the liver model and background skeletal structures, and I’m unsure how to remove the bone background. When using the Segment Editor extension, clicking the 3D Viewer shows blank images instead. Could you please guide me on how to resolve these issues?
Hi Hui Li,
Thank you for your feedbacks.
For 3D Slicer related questions, including usage and support for extensions, it’s best to address them in the 3D Slicer’s discourse.
If you are using the segmentation from the module, then the default display is in MIP (Maximum Intensity Projection).
The projection mode can be changed in the Volume Rendering module in the advanced / techniques options.
Please refer to the module’s documentation if you need more information on this.
If you only want to use the liver segmentation and not the full annotation pipeline, you can directly access it in the segment editor module.
In the segment editor module, no changes to the view layouts is applied.
Best,
Thibault