3D Slicer Resamples Ultrasound Images
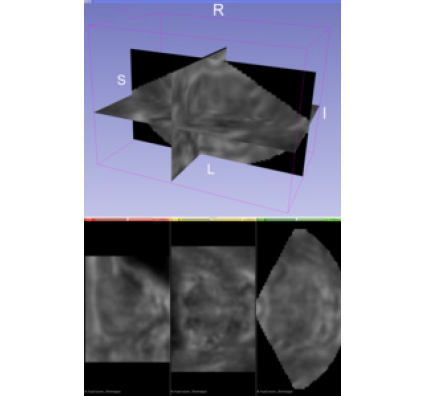
As a medical imaging modality, one of greatest strengths of ultrasound is its ability to inspect the body’s condition in real time. For decades, radiologists, physicians, and ultrasonographers have leveraged traditional two-dimensional (2D) brightness-mode (B-mode) ultrasound images to obtain real-time structural information about soft tissue planes. New opportunities to improve patient care come from imaging disease-specific tissue properties in three dimensions. In particular, opportunities exist in identifying clinically significant prostate cancer lesions by generating three-dimensional (3D) acoustic radiation force impulse (ARFI) images of prostate biopsy candidates [1]. ARFI imaging monitors the mechanical response of tissue to a high-intensity ultrasound-pushing pulse. The process results in an image that reflects local tissue viscoelasticity. Stiffer regions become associated with cancer, whereas less-stiff regions become associated with normal tissue or benign prostatic hyperplasia.
A 3D ARFI image visualizes structural relationships, enables comparison with images from other modalities like multi-parametric magnetic resonance imaging, and ensures the identification of focal lesions. Yet, new computational challenges arise with novel ultrasound imaging methods that deal with three dimensions. Advanced image formation methods like ARFI require more processing than methods like B-mode, and the additional image dimension adds to the computational complexity.
This article describes how the 3D medical image processing, parallel computing, and algorithmic architectures of the Insight Segmentation and Registration Toolkit (ITK) and the Visualization Toolkit (VTK) became integrated into a 3D Slicer extension for ultrasound scan conversion, called SlicerITKUltrasound.
The algorithmic foundation for the SlicerITKUltrasound extension modules [2] is an ITK module, called ITKUltrasound [3]. This module gets built during the extension build process to easily extend the implementation of ITK in 3D Slicer. As an ITK module, ITKUltrasound can easily find use in other contexts outside of 3D Slicer.
Scan conversion resamples processed ultrasound data from its natural transducer sampling geometry onto a uniform grid for visualization and further analysis. Scan conversion is fundamental to all ultrasound imaging modalities, and it must be fast for real-time imaging. First, this article describes the transducer probe geometries that 3D Slicer supports along with their parameterizations. Next, the article highlights resampling algorithms (nine in total). Finally, it discusses the trade-offs and behavioral differences of the resampling algorithms and their implementations.
Geometries
The shape of the transducer probe array determines the ultrasound sampling geometry. A curvilinear array, which is common to abdominal imaging, or a phased array, which is common to cardiac imaging, results in the iconic 2D ultrasound sector image, for example.
3D Slicer modules support three classes of probe geometries. These modules are as follows:
- ScanConvertCurvilinearArray;
- ScanConvertPhasedArray3D; and
- ScanConvertSliceSeries.
ScanConvertCurvilinearArray scan converts ultrasound probes that position elements along a circular arc. This type of probe is common in abdominal imaging. The angular separation between each lateral line, the physical distances between samples along a radial line, and the radius of the arc parameterizes the geometry. ScanConvertCurvilinearArray scan converts additional datasets in the third dimension as equally spaced orthogonal slices.
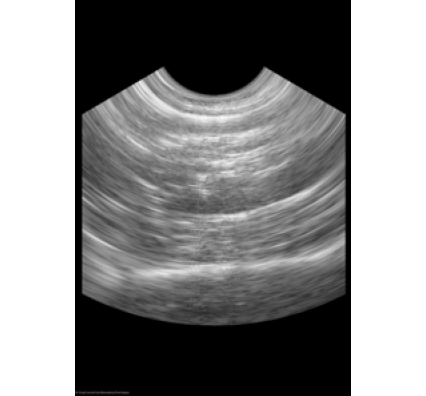
ScanConvertPhasedArray3D scan converts ultrasound probes that position tightly packed elements on a small surface. This type of probe is common in cardiac imaging. When a wave front becomes electrically steered from the surface during the imaging process, it stimulates the transducer elements. The image that results contains a section of a sphere that centers on the probe. The angular separation between each lateral line in the azimuthal direction and the elevational direction, the physical distances between samples along a radial line, and the distance to the first radial sample parameterizes the geometry.
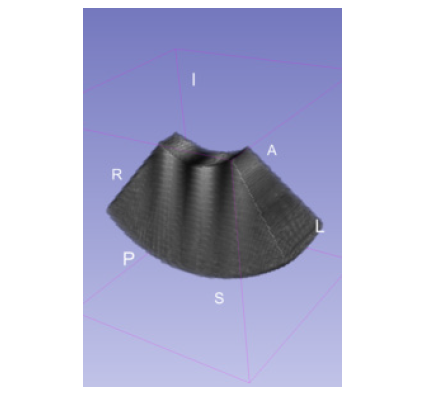
ScanConvertSliceSeries scan converts a set of adjacent, non-overlapping 2D slices. Each slice can have either a curvilinear array geometry or a linear array geometry. These geometries are common with mechanically swept 2D probes, where the distance and the orientation of the intermediate slices may or may not be uniform. In such cases, the geometries of the component slices determine the geometry of the volume.

When a module scan converts a volume, the volume gets resampled onto an itk::Image geometry. This geometry is rectilinear, and it potentially has anisotropic spacing, a physical offset (an origin), and an orientation. This geometry is also the natural geometry for ultrasound probes that position elements along a line, e.g., a linear array.
Resampling Methods
There are three classes of resampling methods. One class, ITK based resampling methods, uses interpolation functions that operate on input samples in the neighborhood of the desired physical output point. The locations of neighborhood input samples pertain to the mapping from output index space to physical space and from physical space to input continuous index space.
The interpolation method sets the number of samples. ITKLinear (the default method) applies trilinear interpolation. ITKNearestNeighbor uses the value of the closest input point to determine the output value. ITKGaussian performs multiplication with a finite-domain Gaussian kernel. Finally, ITKWindowedSinc applies Sinc interpolation with a three-sample radius Lanczos window.
Another two classes operate on a representation of the input as a vtkStructuredGrid data structure. Locator methods that utilize cell topology determine input points. One of these classes, VTKProbeFilter, contains a method that produces an output point, which derives from a weighted combination of points in the surrounding cell.
The other class stems from the new VTK point interpolation framework [4]. While the framework can use a fixed number of nearby points, its implementation for this article considers all the points that fall within a fixed physical radius of an output point. An output point derives from a weighted combination of input points, which the kernel type determines. Methods in the class include VTKVoronoiKernel, VTKLinearKernel, VTKShepardKernel, and VTKGaussianKernel. VTKVoronoiKernel uses nearest-neighbor-like interpolation; VTKLinearKernel employs linear averaging; VTKShepardKernel implements 1 / r^2 weighting; and VTKGaussianKernel utilizes Gaussian weighting.
Discussion
Modules in the SlicerITKUltrasound extension currently enable scan conversion of commonly encountered 2D and 3D ultrasound probe geometries. The extension applies not only to B-mode imaging but to ARFI, ultrasound spectroscopy, and other imaging modes. The performant parallelism in ITK and VTK enable high-throughput computing on emerging many-core computing architectures.
A number of different resampling methods exists. These methods present trade-offs in speed, interpolation errors, and interpolation characteristics. The default resampling method, ITKLinear, offers a good combination of speed and smoothly varying output. Alternatively, ITKWindowedSinc provides the most accurate interpolation method. The violation of the rectilinear input assumption in ITK based methods, however, results in some interpolation errors.
Certain cases, such as the interpolation of segmentation labels, call for the retention of input values. ITKNearestNeighbor or VTKVoronoiKernel is appropriate in these cases. Moreover, with the application of VTKGaussianKernel and ITKGaussian methods, it becomes possible to simultaneously smooth the input and perform interpolation. The physical radius-based interpolation domain of VTK point interpolation methods ensures that consistent spatial content determines an output point. A physical radius-based domain only works well when the spacing of input points is roughly uniform throughout the volume. Otherwise, a computationally taxing number of points in the interpolation domain may result, or the interpolation domain may contain no points at all.
Acknowledgement
Research reported in this publication was supported by the National Institute Of Biomedical Imaging and Bioengineering and the National Institute of General Medical Sciences of the National Institutes of Health under award number R01EB021396 and by the National Cancer Institute of the National Institutes of Health under award number R41CA196565. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
[1] Palmeri, Mark, Tyler Glass, Rajan Gupta, Matt McCormick, Alison Brown, Thomas Polascik, Stephen Rosenzweig, Andrew Buck, and Kathy Nightingale. In 2016 IEEE International Ultrasonics Symposium (IUS). Proceedings of IUS, France, Tours. Institute of Electrical and Electronics Engineers, 2016. http://dx.doi.org/10.1109/ULTSYM.2016.7728618.
[2] “SlicerITKUltrasound.” https://kitwaremedical.github.io/SlicerITKUltrasoundDoc.
[3] KitwareMedical/ITKUltrasound. October 18, 2016. https://github.com/KitwareMedical/ITKUltrasound.
[4] Schroeder, Will. “Point and Smoothed-Particle Hydrodynamics (SPH) Interpolation in VTK” Kitware blog, August 13, 2016. https://blog.kitware.com/point-and-smoothed-particle-hydrodynamics-sph-interpolation-in-vtk.
 Matt McCormick, a technical expert at Kitware, is a major contributor to and the current maintainer of ITK. His general interests include image processing and emphasize scientific reproducibility.
Matt McCormick, a technical expert at Kitware, is a major contributor to and the current maintainer of ITK. His general interests include image processing and emphasize scientific reproducibility.
 Will Schroeder is president and CEO of Kitware. He identifies technology and business opportunities, and he obtains the necessary support for Kitware to meet these opportunities. Schroeder also provides technical leadership for large open-source projects such as ITK and VTK.
Will Schroeder is president and CEO of Kitware. He identifies technology and business opportunities, and he obtains the necessary support for Kitware to meet these opportunities. Schroeder also provides technical leadership for large open-source projects such as ITK and VTK.
 Mark Palmeri is an associate professor of the practice in biomedical engineering at Duke University. His research interests include acoustic radiation force shear wave speed (SWS), ultrasonic imaging, finite element analysis of soft tissue response to acoustic radiation force excitation, medical image processing, and medical instrumentation design.
Mark Palmeri is an associate professor of the practice in biomedical engineering at Duke University. His research interests include acoustic radiation force shear wave speed (SWS), ultrasonic imaging, finite element analysis of soft tissue response to acoustic radiation force excitation, medical image processing, and medical instrumentation design.
 Stephen Aylward is the senior director of operations and the founder of the Kitware office in North Carolina. He helps direct medical research activities at the company. In general, Aylward’s research focuses on vascular image analysis, computer-aided diagnosis, and intra-operative registration of ultrasound images with pre-operative images. He also works to develop ITK and other open-source toolkits.
Stephen Aylward is the senior director of operations and the founder of the Kitware office in North Carolina. He helps direct medical research activities at the company. In general, Aylward’s research focuses on vascular image analysis, computer-aided diagnosis, and intra-operative registration of ultrasound images with pre-operative images. He also works to develop ITK and other open-source toolkits.