3D Slicer and Virtual Insect Dissection
Insects are arguably the most successful terrestrial life form. With an evolutionary history stretching over 400 million years, the world’s oceans remain the sole domain without a significant insect presence. Numbering somewhere around 1.5 million species, their vast diversity remains largely unparalleled.
Scientists have been studying the structure, or morphology, of insects for hundreds of years. The overwhelming majority of studies, however, concentrate only on external anatomy. Investigation of internal structures has usually required one of two techniques: dissection, which distorts the position of internal organs by cutting into and prying open the exoskeleton; or serial sectioning, an extremely labor-intensive process where an insect is embedded in clear epoxy and thinly sliced, one section at a time, to show internal anatomy.
New imaging modalities, in particular micro-CT scanning, have ushered in a renaissance in insect morphology. Here at the American Museum of Natural History in New York, our micro-CT scanner in the museum’s interdisciplinary Microscopy and Imaging Facility allows us to peer into areas previously unstudied.
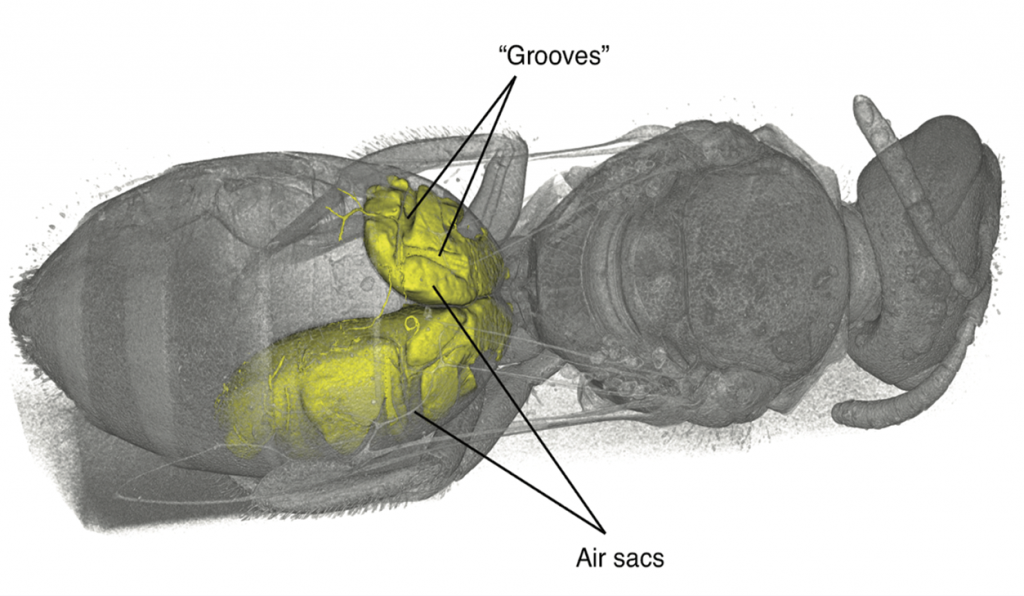
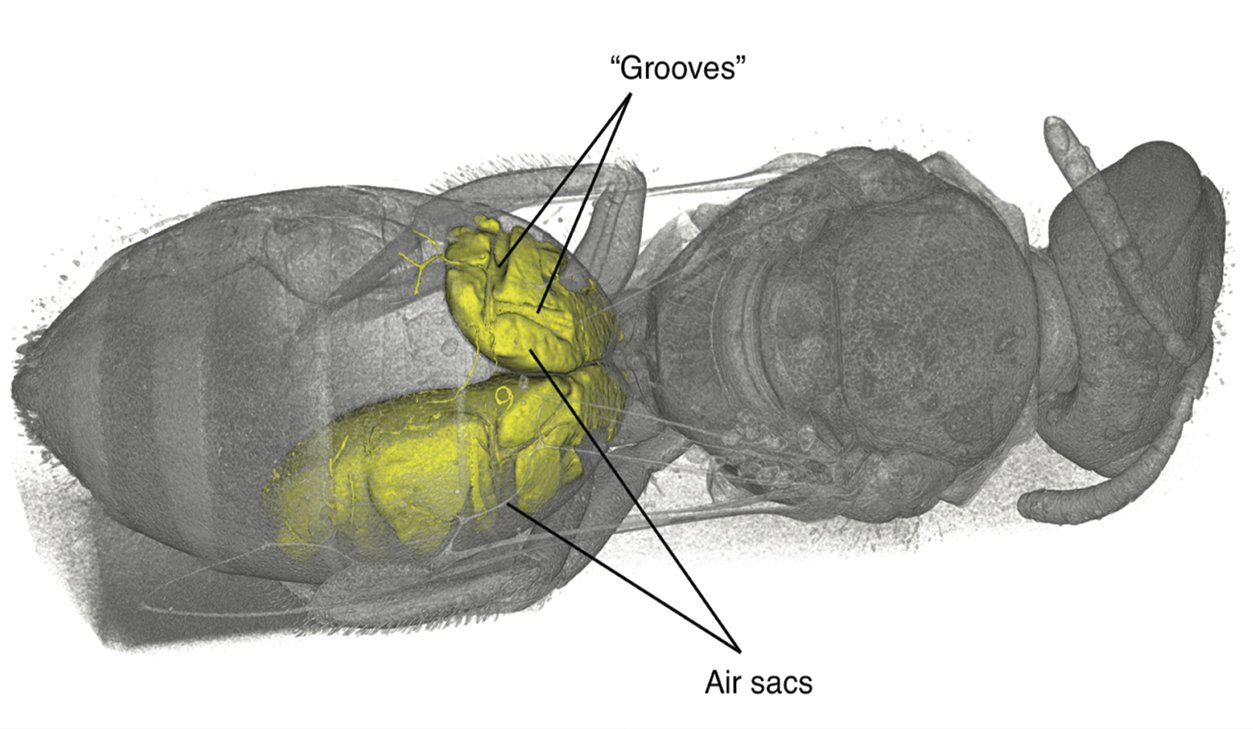
During an investigation of bee anatomy, we came across some unexpectedly large muscles in the abdomens of some “sweat bees,” so called because of their habit of occasionally drinking sweat off desert hikers. Muscles are found in the abdomens, the typically bulbous hind ends, of nearly all insects. These muscles are usually thin and flat, arranged along the inside of the body wall, and are used to pull segments together for abdominal pumping—basically, what looks like insect breathing. What was seen in these bees, however, were large, cylindrical muscles, arranged to pull top-to-bottom—a structure often seen in the thorax (the middle part where the legs and wings attach) for powering the wings, except these were in the abdomen. The actual purpose of these newly discovered muscles is a subject of ongoing research.
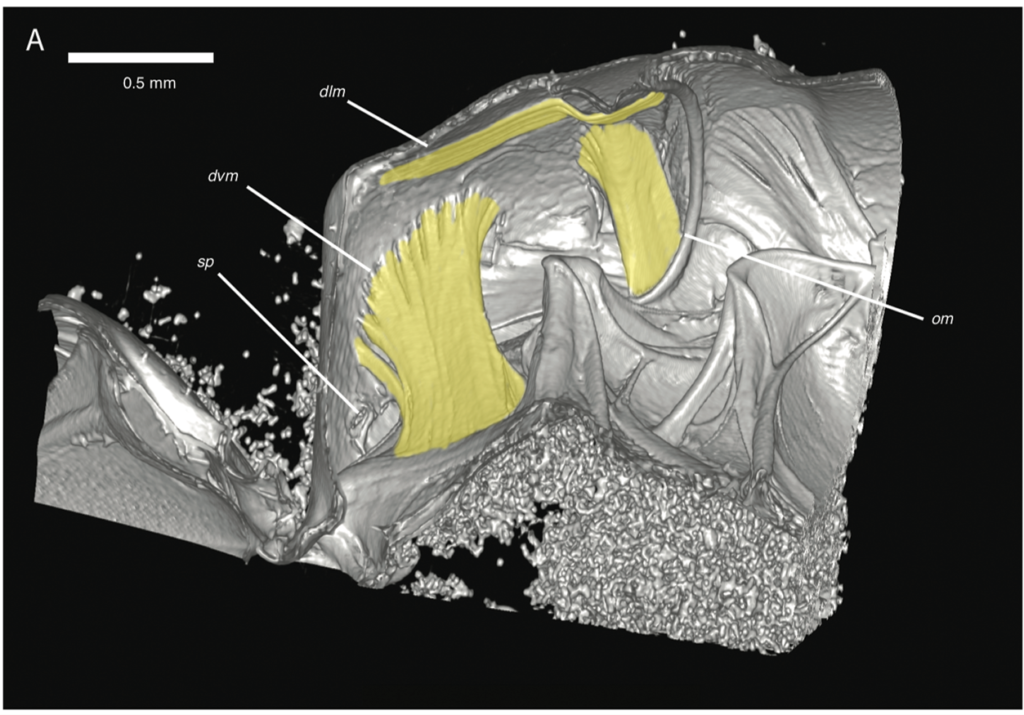
High-resolution, volumetric datasets of insect internal structures are only as useful as the tools used for analysis. Voxel sizes for insect scanning are quite a bit smaller than the typical human-sized scan; for example, the 16 species scanned for the bee study ranged from 4.8 microns to 19.4 microns. We have been using 3D Slicer in our analysis pipeline since 2016. In addition to its vast array of packaged functionality, we have taken advantage of the Python scripting capabilities of 3D Slicer to develop custom analysis tools for various studies, such as area and volume measurement and plotting.
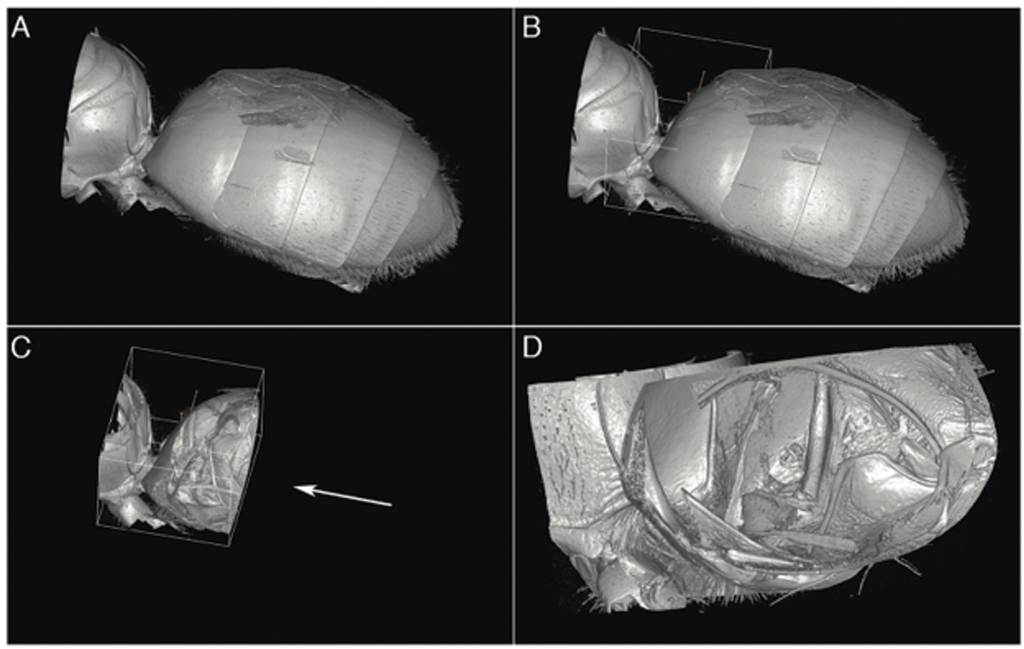
Tube removal is an essential yet simple technique. Insects are typically placed in a plastic Eppendorf tube for scanning. Scan data includes the sample holder as well as the specimen. Removal of the tube is necessary to successfully volume-render the insect, otherwise the result is a great-looking, high-resolution image of an Eppendorf tube. Masking and cropping of the dataset is done using a “fill between slices” technique, where the desired sample area is marked at representative slices spaced throughout the volume along the head-to-tail axis of the insect. Approximately 20-40 slices are marked, depending on the complexity of the specimen and its proximity to the inside surface of the vial. The masking volume is then created using the “Fill between slices” feature of the “Segment Editor Extra Effects” module. Any required cleanup of this volume is done using the standard Segment Editor tools such as Paint and Scissors. Creating this masking volume for tube removal typically takes about an hour, depending on the specimen.
When this masking volume is applied against the original volume with the “Mask Volume” feature, the tube is removed, leaving only the insect. At this point, the masking segmentation is saved as a separate segmentation file to be used later in case missing areas in the cropped volume are noticed during analysis and need to be added back.
Once the volume is masked, standard volume-rendering techniques are used to view the specimen. For the sweat bee study, enabling Cropping and moving the Crop ROI around allowed real-time virtual dissection for detailed examination of internal structures.
A compelling reason for our use of 3D Slicer is interoperability. The cross-platform and open-source nature of the project means data sharing with colleagues is simple. Platform and file format issues are simply nonexistent, as anyone can download and install on any system to view and contribute.
One particular feature seldom highlighted in product marketing is the user community. The Slicer forum is a tremendous resource for users old and new, providing invaluable assistance with using the program, or development assistance for those wishing to create their own extensions and modules.
Insects provide an amazing resource for the study of evolution, and continued improvements in micro-CT scanning and analysis tools will help us learn more about these fascinating animals.
References
[1] Herhold, Hollister W., Steven R. Davis, Corey S. Smith, Michael S. Engel, and David A. Grimaldi. 2019. “Unique metasomal musculature in sweat bees (Hymenoptera, Apoidea, Halictidae) revealed by micro-CT scanning.” American Museum Novitates 2019, no. 3920 (February): 1-28. https://doi.org/10.1206/3920.1

Hollister Herhold is a Visiting Scientist at the American Museum of Natural History, working in the Entomology and Paleoentomological Collections. His research includes studying evolution via insects (fossils as well as living insects and other arthropods), primarily using micro-CT to study anatomy and physiology.